
ID:dan_1622 |
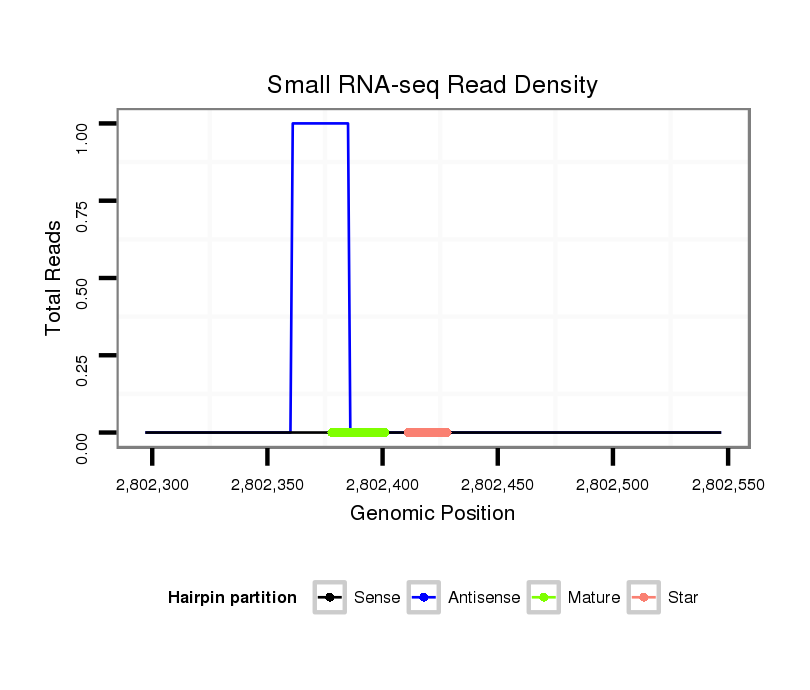
Coordinate:scaffold_13250:2802347-2802497 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -12.0 | -11.3 | -11.3 | -11.1 |
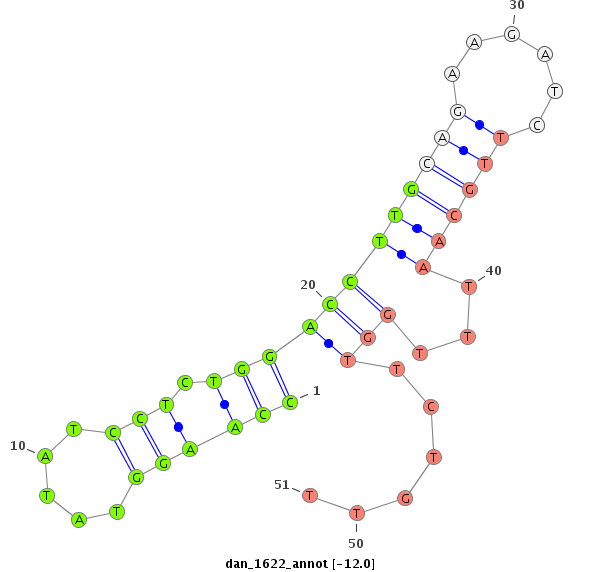 |
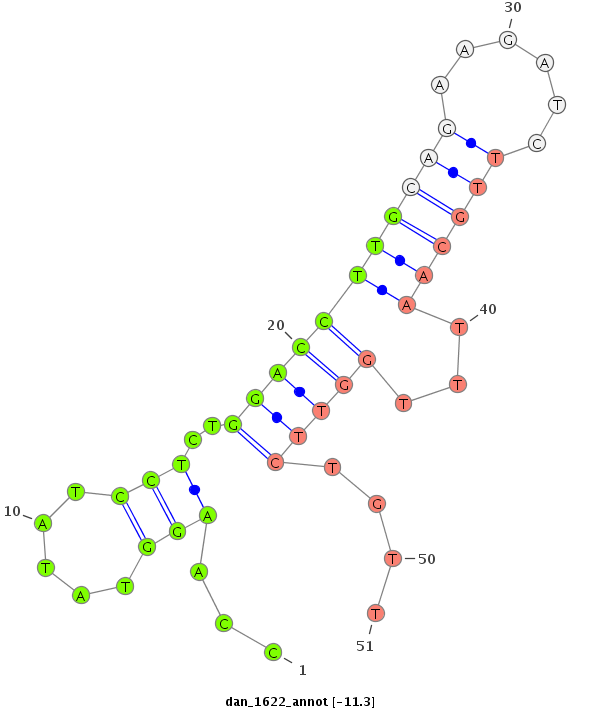 |
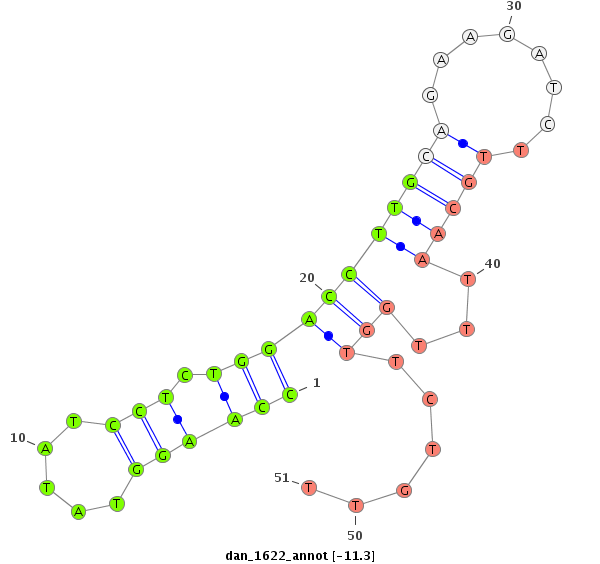 |
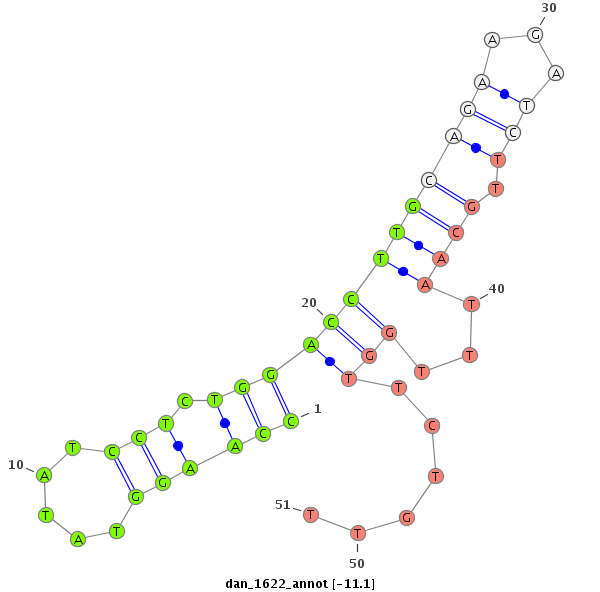 |
exon [dana_GLEANR_1274:2]; CDS [Dana\GF12722-cds]; intron [Dana\GF12722-in]
| Name | Class | Family | Strand |
| OSVALDO_I-int | LTR | Gypsy | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTAAATTTATTTTGATGATGTGTTATATGTATACATGAAAAGAACCACCCCCGTCCGCCCTTCACTCCCATCCTTACGTCCCCAAGGTATATCCTCTGGACCTTGCAGAAGATCTTGCAATTTGGTTCTGTTTTAACAACAGCTCGAACTACACACATTACAGTAGCATTACAGTATCAGTATAAAGTATCATCCATTCAGGGAAGAGCTGCTGTACTTGGGTCACCGAATAAAAAGCCAGGGAATCGGCA *********************************************************************************..((............(((((((((((......))))))...)))))..))*********************************************************************************************************************** |
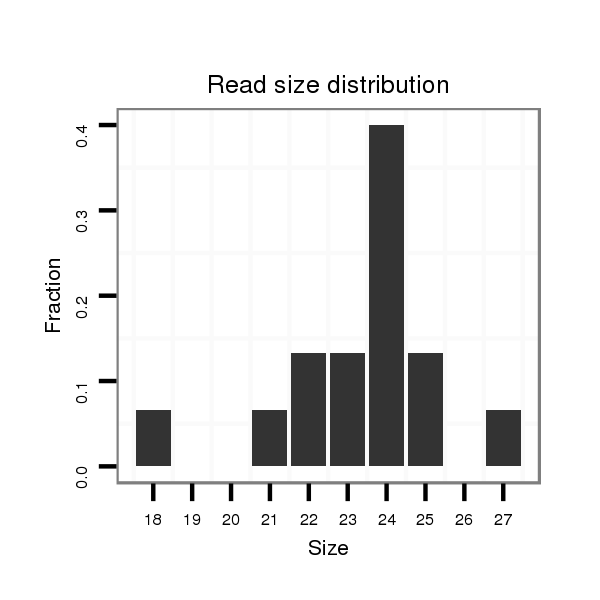
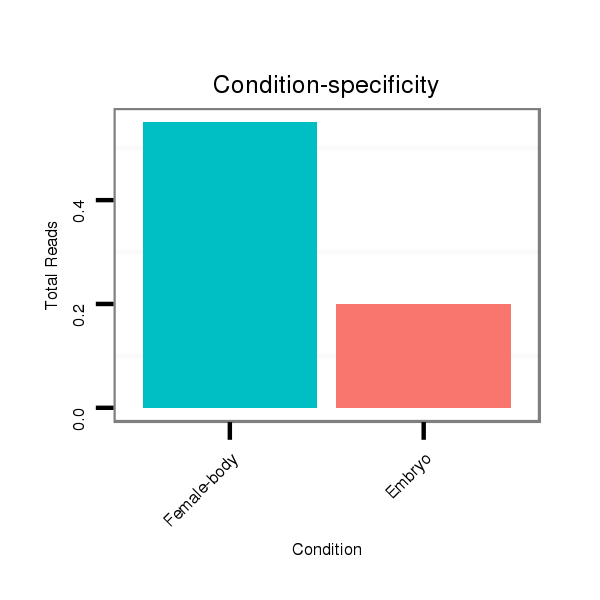
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V039 embryo |
M044 female body |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................CATCTAATCAGGGAAGAGC.......................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..........................ATGTCTACATGAGAGGAACCAC........................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................ATGTCTACATGAACAGCACCA............................................................................................................................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................AAAATACAGGGAATCGGCA | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 |
| .................................................................................CCAAGGTATATCCTCTGGACCTTG.................................................................................................................................................. | 24 | 0 | 20 | 0.15 | 3 | 0 | 0 | 3 |
| .................................................................................CCAAGGTATATCCTCTGGACCTT................................................................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 |
| ..................................................................................CAAGGTATATCCTCTGGACCTTGC................................................................................................................................................. | 24 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 |
| ...............................................................................................................................................................................................................GCGGCTGTACGTGGGGCACC........................ | 20 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 |
| ......................................................................................GGATATCCTCTGGACCTTGC................................................................................................................................................. | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .................................................................................CCAAGGTCTATCCTCTGGACCA.................................................................................................................................................... | 22 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...............................................................................CCCCATGGTATATCCTCTGGACCTT................................................................................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..................................................................................CAAGGTATATCCTCTGGACCTTGCA................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ...................................................................................AAGGTATATCCTCTGGACCTTG.................................................................................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ...........................................................................ACGTCCCCAAGGTATATCCTC........................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..................................................................................................................TTGCAATTTGGTTCTGTT....................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...............................................................................CCCCAAGGTATATCCTCTGGACCTTGC................................................................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| .........................................................................................TATCCTCTGGACCTTGAAA............................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...............................................................................CCCCAAGGTATATCCTCTGGAC...................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..............................................................................TCCCCAAGCTATATCCTCTGGACCTTGC................................................................................................................................................. | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..............................................................................TCCCCAAGGTATATCCTCTGGACCT.................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ...............................................................................CCCCAAGGTATATCCTCTGGACCT.................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
AATTTAAATAAAACTACTACACAATATACATATGTACTTTTCTTGGTGGGGGCAGGCGGGAAGTGAGGGTAGGAATGCAGGGGTTCCATATAGGAGACCTGGAACGTCTTCTAGAACGTTAAACCAAGACAAAATTGTTGTCGAGCTTGATGTGTGTAATGTCATCGTAATGTCATAGTCATATTTCATAGTAGGTAAGTCCCTTCTCGACGACATGAACCCAGTGGCTTATTTTTCGGTCCCTTAGCCGT
***********************************************************************************************************************..((............(((((((((((......))))))...)))))..))********************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V055 head |
M058 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|
| ...................................CCTTTTCATCGTGGGGGCAGG................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................................................GAGGGTAGGAATGCAGGGGTTCCAT.................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................ATAGGAGACCTGGAACGTCTTCTAGAAC...................................................................................................................................... | 28 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................AGGAGACCTGGAACGTCTTCTAGAAC...................................................................................................................................... | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................ATTCATAGTAGGTGAGTCC................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ...........................................TGGTGGGGGCAGACGTGAGGTG.......................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................CAAGAAGAAATTGTTGTC............................................................................................................. | 18 | 2 | 17 | 0.35 | 6 | 1 | 4 | 0 | 1 |
| ...........................................................................................AGGACACCTGGAACGTCTTCTAGAAC...................................................................................................................................... | 26 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .........................................CTTCGTGGGGGCAGGCGGC............................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ..........................................TTGGTGAGGGTAGGCAGGAAG............................................................................................................................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .........................................................GGGAAGTGAGCGTAGCAA................................................................................................................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 |
| .............................................GTGGGGGCAGGCGGGG.............................................................................................................................................................................................. | 16 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................GTGAGGGTAGTAATGGAG........................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
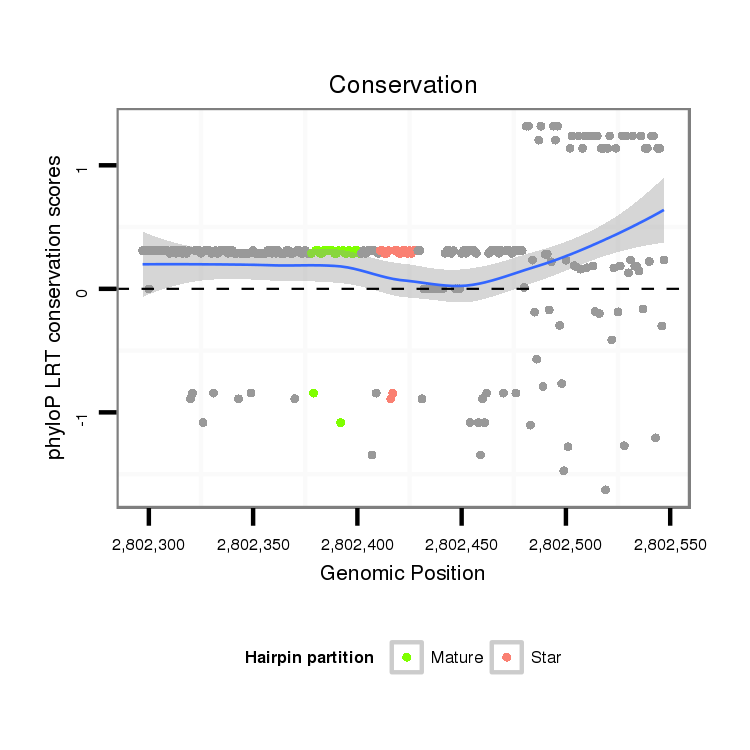
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13250:2802297-2802547 + | dan_1622 | TTAAATTTATTTTGATGATGTGTTATATGTATACATGAA-AAGAACCACCCCCGTCC---------------------------------GCCCTTCACTCCCATCCTTACGTCCCCAAGGTATATCCTCTGGACCTTGCAGAAGATCTTGCAATTTGGTTCTGTTTTAACAACAGCTCGAACTACACACATTACAGTAGCATTACAGTAT-------CAGTATAAAGTATCATCCATTCAGGGAAGAGCTGCTGTACTTGGGTCACCGAATAAAAAGCCAGGGAATCGGCA |
| droBip1 | scf7180000396583:97730-97956 - | TTA-ATTTATTTTGATGATGTGTAGTATGGATACGTGAAAAAGAACCTCCCCCATCCGTCGAGCTAAGTCTAAGACCACAAGAAGTGCACGCCCTTCACTCCCATCCATACGTCCCTAAGGTATATCCTATGGACCTTGCAGAACACCTTGCATCTTGGTTCTGTTTTT----------GAACT---CACAGTACCCACACATTACAATATTTGGGCCCAATATGAAA-GTCAGCCAGTCAG-------------------------------------------------- | |
| droFic1 | scf7180000454097:334-400 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATGCCAGTTCTTCAGGCAAGAGTTGCTGTACCTAGGACACCGAGTAACAAGCCAAGGGATAGGGA | |
| droEle1 | scf7180000486270:8688-8755 + | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAGAGTCATTTCTTCAGAAGCGAGCTGCTTTACCTGGGGCACCGCGTTACAAATCAAGGAATCGGCT | |
| droRho1 | scf7180000774314:110-176 - | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGTCAGTTCTTCAAGCAGGAGCTGCTGTACCTGGGACATCGAGTAACAAGCCAAGGAATCGGTA | |
| droBia1 | scf7180000301342:42577-42644 + | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATACCAGTTTTTCAAGAAGGAGCTGCTGTATTTGGGTCATAGAGTGACTAGCGACGGAATAGGCA | |
| droTak1 | scf7180000412828:9523-9589 - | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGTCAATTCTTCAAAAAGGAGCTGCTGTACCTGGGACATCGAGTAACAAGCCAAGGAATTGGCA | |
| droYak3 | v2_chr2L_random_011:34617-34684 + | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATGCCAGTTCTTCAAACAAGAACTTCTGTACCTAGGGCACCGTGTCACAAGCCAAGGAATAGGCA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/18/2015 at 02:33 AM