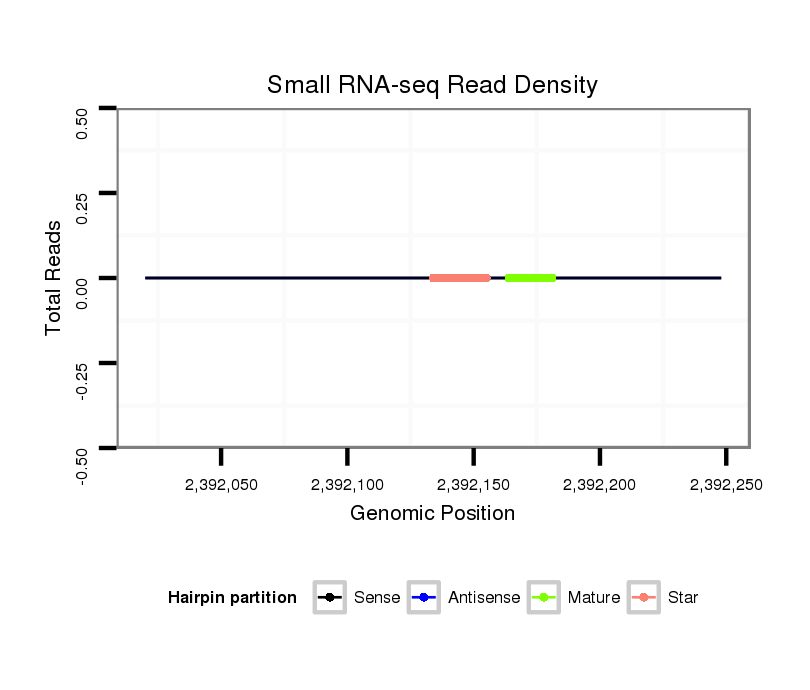
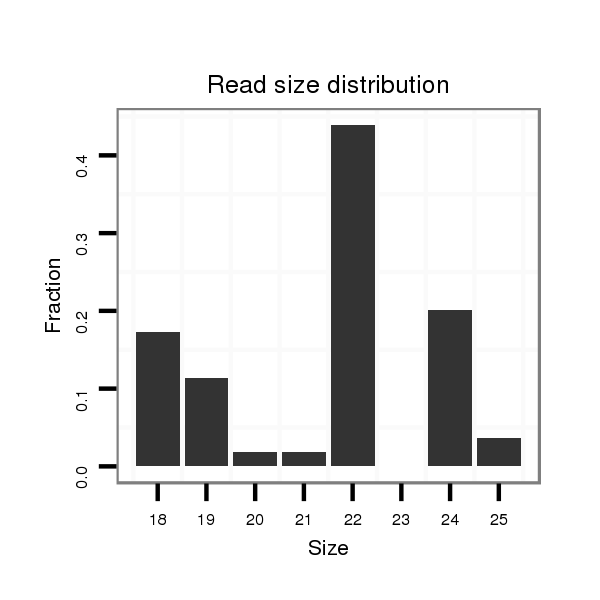
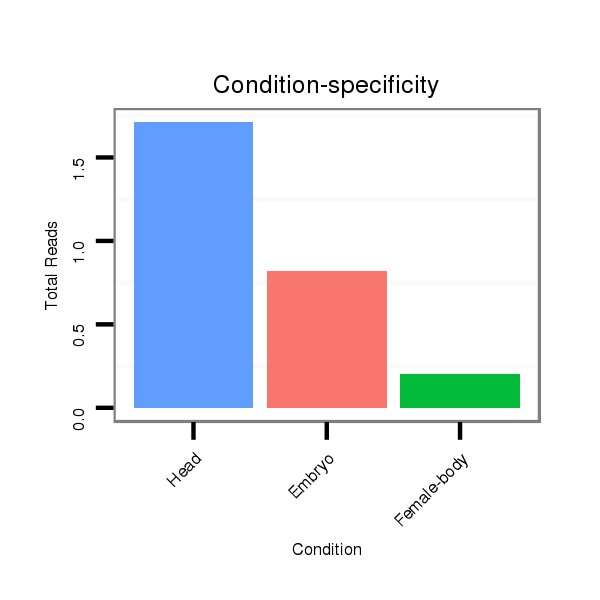
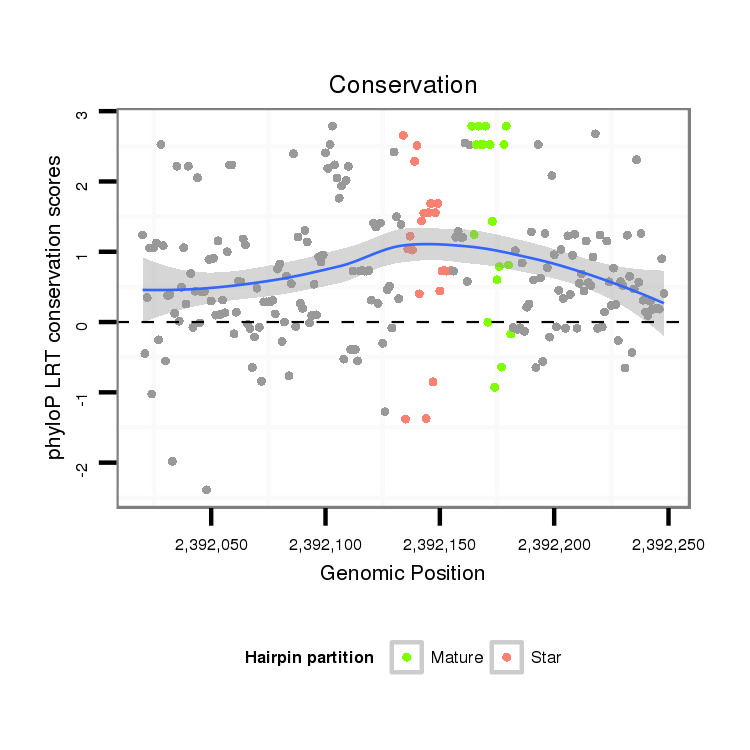
| CACTTCTG--ACCGAGACTTTTGAGGCATTCAACATTTGTCGCTTGT----CCATTG---------------------------------------------------CAGA----------TGGA----AACGCAGA----CAA--CG----CAGCA-GCAACA---------------GCAGCAGCAGCAGCAGCAACAA-----T---TGCAACA---GCAGCAACATCA---------A-----------CAGCAACAACAG------CAGC---------------------AGCAGCAGCAACAACAGCA------------------------ACAGCAGCAACAACAG------CAGCAGGTCCAGGTCCTGACCAAGCAGCAG-----ATCCCATTGATAGACAAGAT-----------------------CAA--GCTAGAGGCC--------------------------------- | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................GATAGACAAGAT-----------------------CAA--GCTAGAGGCC--------------------------------- | 25 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................GGTCCTGACCAAGCAGCAG-----ATCCC...................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................G------CAGCAGGTCCAGGTCCTGAC.......................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................AACAACAG------CAGCAGGTCCAGGTCCT............................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................GACCAAGCAGCAG-----ATCCCATTGATA............................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................ATCCCATTGATAGACAAGAT-----------------------CAA--GCTAGA..................................... | 29 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................CAGCAGGTCCAGGTCCTGACCAAGCAGC.................................................................................................. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................ATCCCATTGATAGACAAGAT-----------------------................................................ | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................AGA----CAA--CG----CAGCA-GCAACA---------------GCA................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................GGTCCAGGTCCTGACCAAGCAGCAG-----........................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GA----------TGGA----AACGCAGA----CAA--CG----CAGC........................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................AGCAACATCA---------A-----------CAGCAACAACAG------CAGC---------------------............................................................................................................................................................................................... | 27 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................GCAGCAACAA-----T---TGCAACA---GC......................................................................................................................................................................................................................................................................... | 20 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................GCAACA---GCAGCAACATCA---------A-----------CA........................................................................................................................................................................................................................................ | 21 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................ATCA---------A-----------CAGCAACAACAG------CAGC---------------------AG............................................................................................................................................................................................. | 23 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................CAACAACAGCA------------------------ACAGCAGCAACAACA..................................................................................................................................... | 26 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................ACAACAG------CAGC---------------------AGCAGCAGC...................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................GCA-GCAACA---------------GCAGCAGCAGCAGCAGCAAC................................................................................................................................................................................................................................................................................................ | 29 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................AGCAACAACAGCA------------------------ACAGCAGC............................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................AGCAGCAACAACAGCA------------------------ACA................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................A-----------CAGCAACAACAG------CAGC---------------------A.............................................................................................................................................................................................. | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................AGCAGCAGCAACAACAGCA------------------------ACAGCA.............................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................AACAACAGCA------------------------ACAGCAGCAAC......................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................CAGCAACAACAG------CAGC---------------------AGCAGCA........................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GC---------------------AGCAGCAGCAACAACAGCA------------------------ACAGCA.............................................................................................................................................. | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................C---------------------AGCAGCAGCAACAACAGCA------------------------ACA................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................AACAACAG------CAGC---------------------AGCAGCAGC...................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................CAACAACAGCA------------------------ACAGCAGCA........................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................ACAACAG------CAGC---------------------AGCAGCAG....................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................AACAACAG------CAGC---------------------AGCAGCAGCA..................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |