
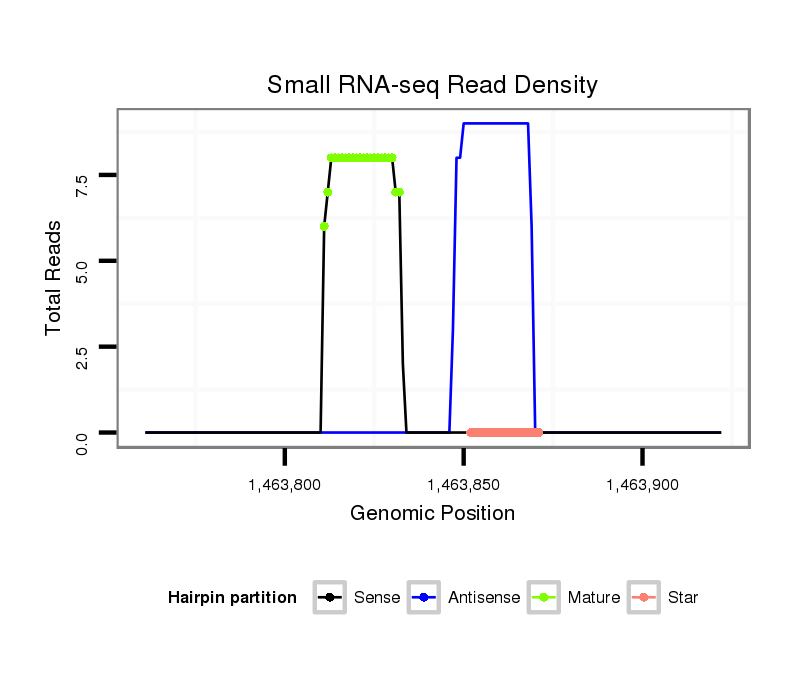

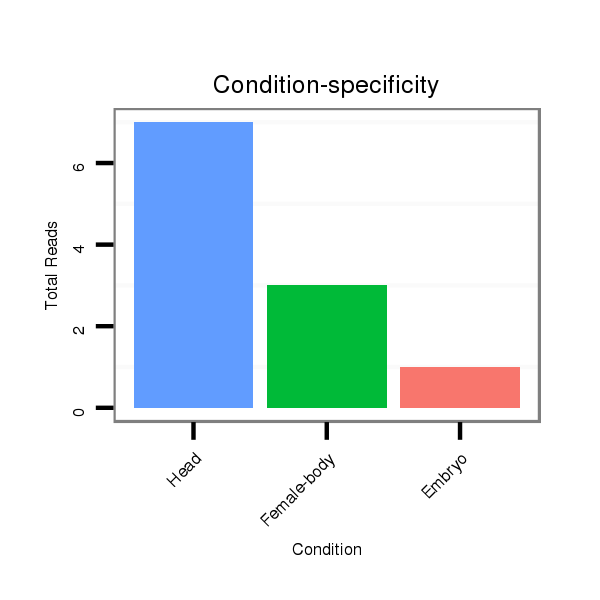
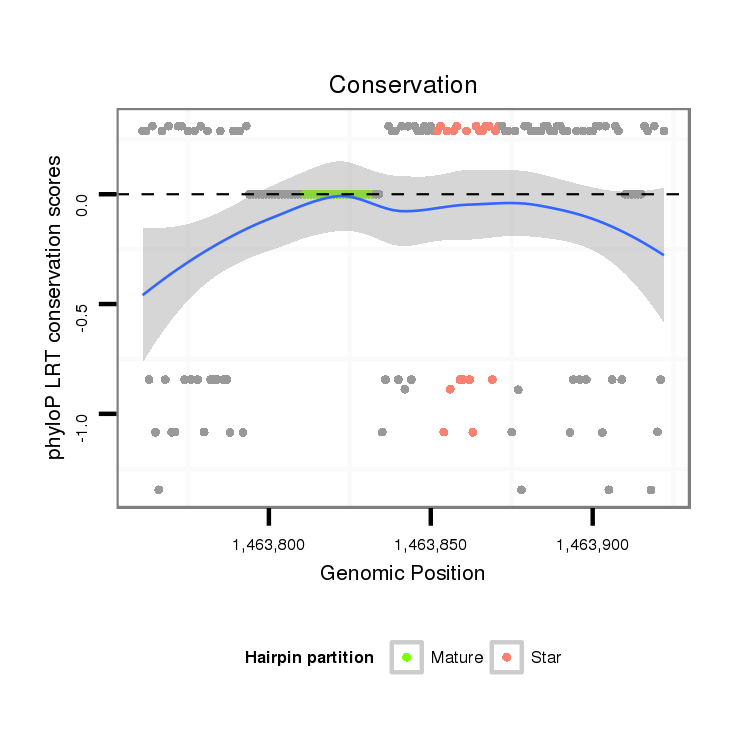
ID:dan_152 |
Coordinate:scaffold_12947:1463811-1463872 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -53.4 | -52.8 |
 |
 |
Antisense to intron [Dana\GF19008-in]
No Repeatable elements found
| mature | star |
|
CCGAAGGTTATTTTGTCAATGCGGCAGCGGGCATCTTCGATCAGTCCATCTTTGGATTCTCTGACAAGGACTTATGACCCTTACTGGAGTCCTTGTCAGAGAATCCAAAGATGGTCAGATCCCCAAGGAACGCTGGGCGGTACAGAAGCCTCGCTTGGAGAG
***********************************...((((...((((((((((((((((((((((((((((.............))))))))))))))))))))))))))))...)))).....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M044 female body |
V105 male body |
M058 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TTTGGATTCTCTGACAAGGACT.......................................................................................... | 22 | 0 | 2 | 5.50 | 11 | 8 | 2 | 0 | 1 | 0 |
| ..................................................TTTGGATTCTCTGACAAGGA............................................................................................ | 20 | 0 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 | 0 |
| ....................................................TGGATTCTCTGACAAGGACTT......................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TTGGATTCTCTGACAAGGACTT......................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................TTGGATTCTCTGACAAGGA............................................................................................ | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................CTTTGGATTCTCTGACAAGGACT.......................................................................................... | 23 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................GGTCAGATCCCGAAGGAAGC.............................. | 20 | 3 | 4 | 0.50 | 2 | 1 | 0 | 1 | 0 | 0 |
| .................................................CTTTGGATTCTCTGACAAGGA............................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TCTGGATTCTCTGACAAGGACT.......................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TTTGGATTCTCTGACAAGG............................................................................................. | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TTTGGATTCTCTGACAGGGACT.......................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TTTGGATTATCTGACAAGGA............................................................................................ | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................TCTTTGGATTCTCTGACAAGG............................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................CTTGTCAGAGAATCCAAAGA................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TTTGGATTCTCGGACAAGGACT.......................................................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................TCTTTGGATTCTCTGACAAGGA............................................................................................ | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................TGGATTCTCTGACAAGGACT.......................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .CGTTGGTGATTTTGTCAATGC............................................................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| CCGTTGGTAATTTTGTCAATG............................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................GGTCAGATCCGGAAGGAA................................ | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................CGAAGATGATCAGATACCCA..................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................GGTCAGATCCGCAAGGAAGC.............................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
|
GGCTTCCAATAAAACAGTTACGCCGTCGCCCGTAGAAGCTAGTCAGGTAGAAACCTAAGAGACTGTTCCTGAATACTGGGAATGACCTCAGGAACAGTCTCTTAGGTTTCTACCAGTCTAGGGGTTCCTTGCGACCCGCCATGTCTTCGGAGCGAACCTCTC
************************************...((((...((((((((((((((((((((((((((((.............))))))))))))))))))))))))))))...)))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M044 female body |
M058 embryo |
V105 male body |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................TCAGGAACAGTCTCTTAGGTTT..................................................... | 22 | 0 | 2 | 5.50 | 11 | 8 | 2 | 1 | 0 | 0 |
| ......................................................................................CTCAGGAACAGTCTCTTAGGTT...................................................... | 22 | 0 | 1 | 3.00 | 3 | 0 | 1 | 2 | 0 | 0 |
| .........................................................................................AGGAACAGTCTCTTAGGTTT..................................................... | 20 | 0 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 | 0 |
| .......................................................................................TCAGGGACAGTCTCTTAGGTTT..................................................... | 22 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................AGGAACAGTCTATTAGGTTT..................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................AGGAACAGTCTCTTAGGTT...................................................... | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................AGGAACAGTCTCTTAGGTTTCT................................................... | 22 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................TCAGGAACAGTCTCTTAGGTTTC.................................................... | 23 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................GGAACAGTCTCTTAGGTTT..................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................TCAGGAACAGTCTCTTAGGT....................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................AGAAACCTAAGAGACTGTTC.............................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................TCAGGAACAGTCTCTTAGGTCT..................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................GGAACAGTCTCTTAGGTTTCT................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................AGGAACAGTCTCTTAGGTTTC.................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................TCAGGAACAGGCTCTTAGGTTT..................................................... | 22 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................TTCCTGAATAGTGGGCATCAC............................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................TTCTACCAGGCTAGGGAT..................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................CAACATGTCTTCGAAGCGA....... | 19 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_12947:1463761-1463922 + | dan_152 | CCGAAGGTTATTTTGTCAATGCGGCAGCGGGCATCTTCGATCAGTCCATCTTTGGATTCTCTGACAAGGACTTATGACCCTTACTGGAGTCCTTGTCAGAGAATCCAAAGATGGTCAGATCCCCAAGGAAC------GCTGGGCGGTACAGAAGCCTCGCTTGGAGAG |
| droBip1 | scf7180000391825:11401-11521 + | CCAACCGCTCGTTCGCCGAGGTAACGAAGGGAA-----------------------------------------GAACCTTAATTGGAGTCCTGGACAAGGGCTCCAAGGATGGGCTCATCCCCAAGGAACTTTGGCGACGAGTGGTAAACGAGT------TGCATGG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 05/15/2015 at 02:55 PM