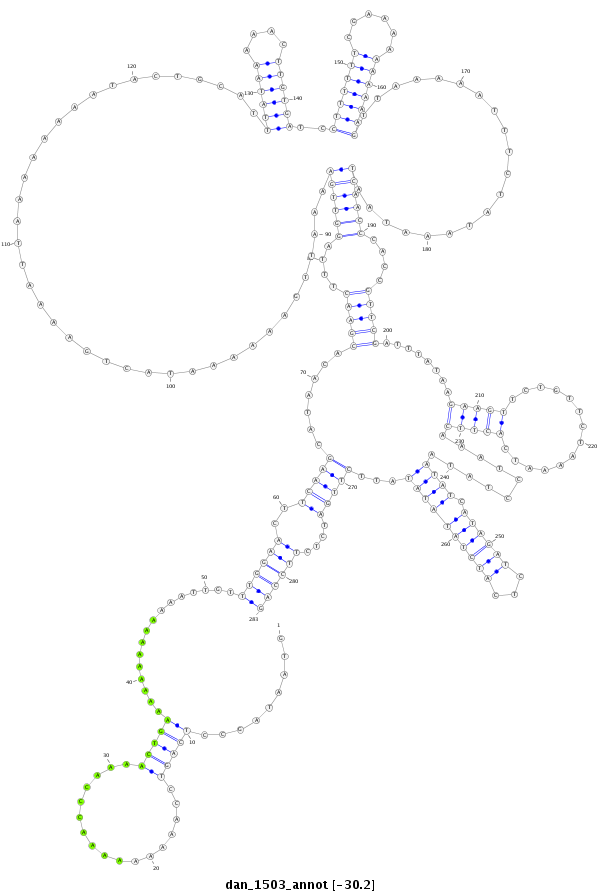
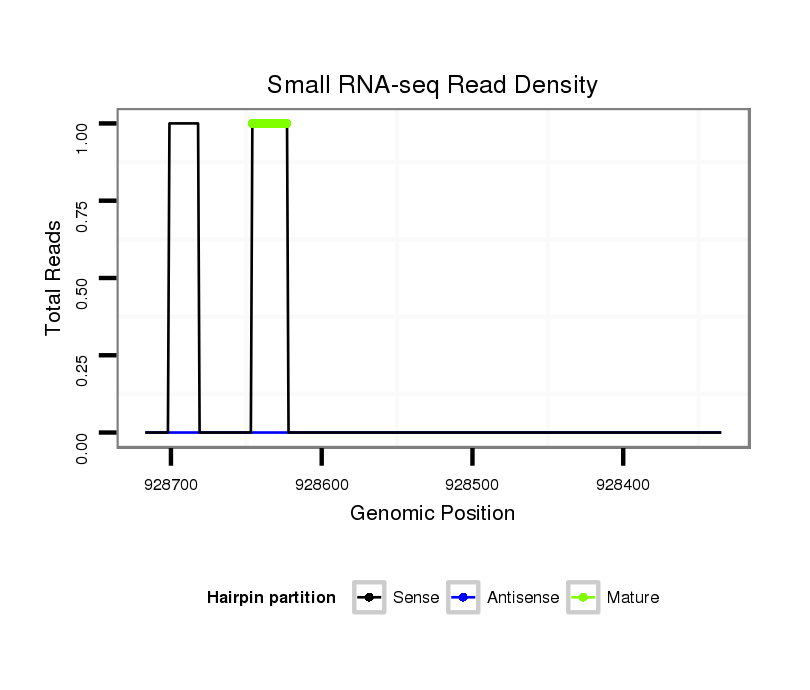
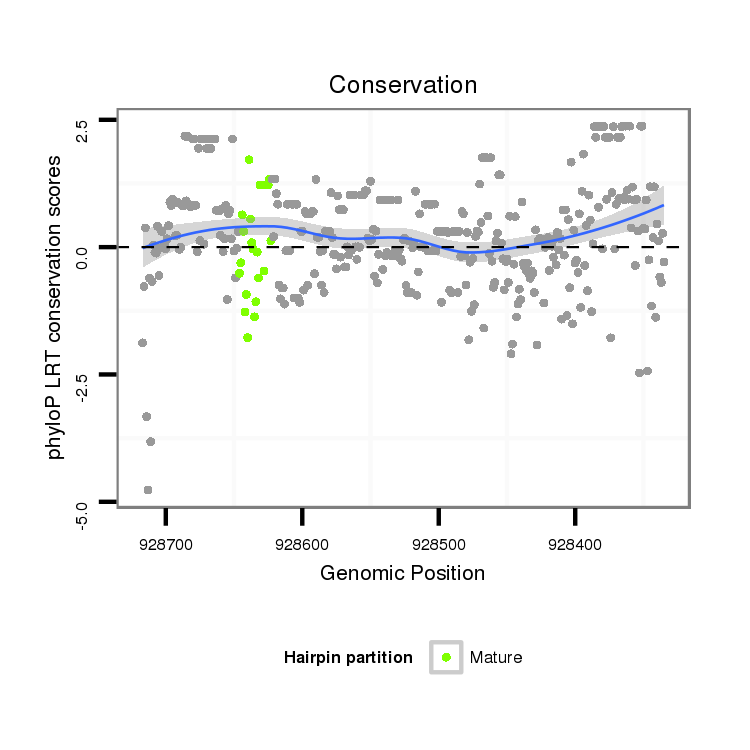
ID:dan_1503 |
Coordinate:scaffold_13248:928385-928667 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -30.2 | -30.2 | -30.2 |
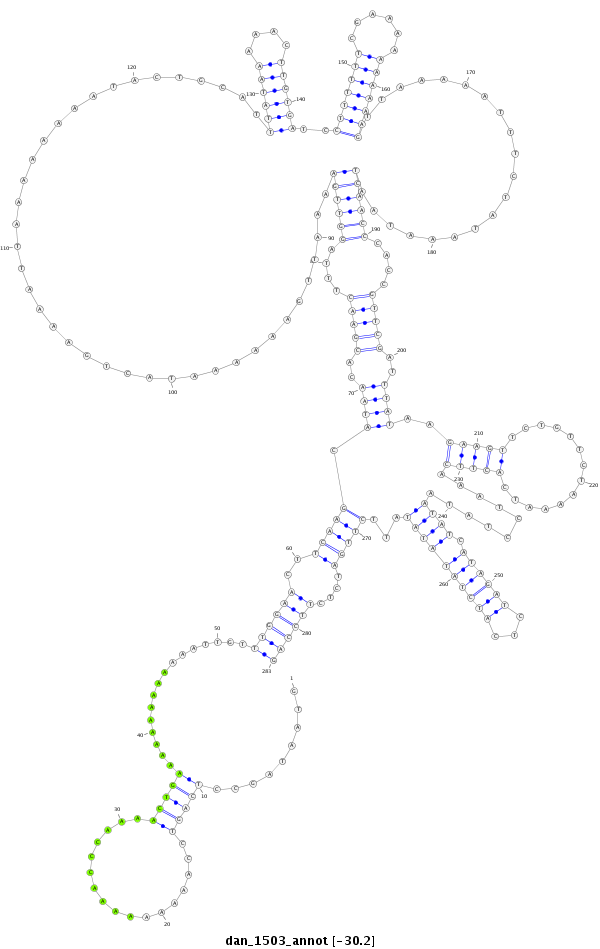 |
 |
 |
CDS [Dana\GF22432-cds]; CDS [Dana\GF22432-cds]; exon [dana_GLEANR_6401:4]; exon [dana_GLEANR_6401:3]; intron [Dana\GF22432-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GATGAACTTGGATCCGAACTACTACCTGACCAATCGGATACCCTATACCAGTAATAGCCTCAGTCCAAAAAAAAACCCAAAACTGAAAAAAAAAAAAATTGTTTGGAACTTCAAGCATAACACGAACTTTAGGTTGAAAATTGAAAAAATACTGAAAATTAAAAAAAATACTGCATTTATAAAAACTTGTGATCCTTTTTTCGAAAAAAAAAAGTTAAAAATTTCTATAAATAATCAACCCACCGTTCGATTTATAAGAAGTTCTGTTCTAAAATCACTTCAAATCCTATAATATCATAGATCTCATCTATATATATTCTTGATCTCTTCCAGACAATAGCGGATTGGCGGAGGATCAGACGCGAAAGAGAGAAATCCGGCTG **************************************************.........(((((.................)))))................((((((..(((((.......(((((....((((((.......................................((((((....))))))..(((((((......)))))))....................))))))....)))))........(((((..............)))))..........((((.((((((...))))))))))...)))))....))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V105 male body |
V055 head |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................AAAACCCAAAACTGAAAAAAAAAA................................................................................................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................GTTGGCGGAGGTTCAGACGC.................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................TTGGCGGAGGTTCAAACGC.................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................ACCAAAAACTGAAAAAAAAAAAAA............................................................................................................................................................................................................................................................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................GCGAAAGAGAGAAATCCGGCC. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................TGACCAATCGGACAACCTATT................................................................................................................................................................................................................................................................................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................AACTACTACCTGACCAATCG........................................................................................................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................AAAAAAAAAAAATTGTTTGGGA................................................................................................................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................TGGCGGAGGTTCAAACGC.................... | 18 | 2 | 4 | 0.75 | 3 | 0 | 3 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................TTGGCGGAGGTTCAAACG..................... | 18 | 2 | 5 | 0.60 | 3 | 0 | 3 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................GTTGGCGGAGGTTCAAACGC.................... | 20 | 3 | 10 | 0.60 | 6 | 0 | 3 | 2 | 0 | 1 |
| ..................................................................AAAAAAAAACCCAAAACGGA......................................................................................................................................................................................................................................................................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................GAATTCACCCACCGTTCGATT................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................CCAAAAAAAAACCCATAAC............................................................................................................................................................................................................................................................................................................ | 19 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................AAAACCCAAAACAGAACAAAAAAAA............................................................................................................................................................................................................................................................................................... | 25 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................CAAACAAACACCCAAAACAGAAAAAA.................................................................................................................................................................................................................................................................................................... | 26 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................AAAGAAACCCAAAACAGAAAAACAAA................................................................................................................................................................................................................................................................................................. | 26 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................TTGGCGGAGGTTCAAACGCC................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................GATTGGGGGAGGATAAGA....................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................AAAAAAGCCAAACCTGAAAA...................................................................................................................................................................................................................................................................................................... | 20 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................AAAAAAAGACCAAACCTGAAAAAA.................................................................................................................................................................................................................................................................................................... | 24 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................AAAACCCAAGACCGAAGAAAAAA................................................................................................................................................................................................................................................................................................. | 23 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................ACCCAACATGGAAAAAAAAAAAAA............................................................................................................................................................................................................................................................................................. | 24 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........AACAAAACTACTACCTGACC................................................................................................................................................................................................................................................................................................................................................................ | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................AAATACACAAAACTGAAAAAAA................................................................................................................................................................................................................................................................................................... | 22 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................AATTGTTTGGATCTTGACGC........................................................................................................................................................................................................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................ATCCTTTTTTACAAAAAAAA............................................................................................................................................................................ | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................ATTGGTGGAGGATCTGAAGC.................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................ATAACACGATCTTGAGTTT........................................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
CTACTTGAACCTAGGCTTGATGATGGACTGGTTAGCCTATGGGATATGGTCATTATCGGAGTCAGGTTTTTTTTTGGGTTTTGACTTTTTTTTTTTTTAACAAACCTTGAAGTTCGTATTGTGCTTGAAATCCAACTTTTAACTTTTTTATGACTTTTAATTTTTTTTATGACGTAAATATTTTTGAACACTAGGAAAAAAGCTTTTTTTTTTCAATTTTTAAAGATATTTATTAGTTGGGTGGCAAGCTAAATATTCTTCAAGACAAGATTTTAGTGAAGTTTAGGATATTATAGTATCTAGAGTAGATATATATAAGAACTAGAGAAGGTCTGTTATCGCCTAACCGCCTCCTAGTCTGCGCTTTCTCTCTTTAGGCCGAC
**************************************************.........(((((.................)))))................((((((..(((((.......(((((....((((((.......................................((((((....))))))..(((((((......)))))))....................))))))....)))))........(((((..............)))))..........((((.((((((...))))))))))...)))))....))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
V039 embryo |
M044 female body |
M058 embryo |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................GTGATTGCAATCCAACTTTGA.................................................................................................................................................................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................CAACCGCCTCCAAGTTTGCG.................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................AGCACTAGGAAAACAGCTCT................................................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TGTTTGAGCACTAGGAAAA........................................................................................................................................................................................ | 19 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AAGAAAGCTTGGTTTTTTCAA....................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................CACCACTAGGAAAAAAGATT.................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............AGGATTGGTGATGGGCTGG................................................................................................................................................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................................................TTAATGATATTTATTAGTTC................................................................................................................................................ | 20 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................GCAGGAAATCCAACTTGTA.................................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AGCACTAGGAAAATAGCTC.................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13248:928335-928717 - | dan_1503 | GATGAACTTGG------ATCCGAACTACTAC---CTGACCAATCGGATACCCTA---TACCAGTAATA--GCCTCAGTCCAAA----------------AAAAAACCCAAAACTGAAAAAAAA-----------------------------------------------------------------------------------------------------AAAAATTGTTTGGAACTTCAAGCATAACACGAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTAGGTTGAA--------------------------------------------------------------------------AATTGAAAAAATACTGAAAATTAAAAA---AAATA-CTGCATTTATAAAAACTTGTGATCCTTTTTTCGAAAAAAAAAAGTTAAAAATTTCTATAAATAATCAACCCACCGTTCGATTTATAA-GAAGTTCTGTTCTAAAAT-CAC---------------------------------------------TTC----A------AATCCTA-----------------------------TAATATC--------------------------------ATAGATCTCAT------C---------TATA-T--ATAT---TCTTG---ATCTCT---------T-------CCAGACAATAGCGGATTGGCGGA-----GGATCAGACGCGAAAGAGAGAAATCCGGCTG |
| droBip1 | scf7180000395920:37611-37962 + | GATCACCATGG------ATCCGAACTACTAC---CTGACCAATCGGATACCCTA---TACCAGTAATA--GATTCAGTCCAAA----------------AA---AATAAAAAAAAGAAAGAAATCTATAAA--------------------------------------------------------------------------------------AATTTAAAAAAATTAAAAAGAACTTTAAAAATGACTCGAAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTAAGATGAA-------------------------------------------------------AT------------------ATTG--------------------AAAAAAAATA-CT-----------------TAATTTTAATTTT-----AAAAAAGTTAAACATTTCCTAAAAAAATCATCTTTTAGTTCGAGTTATAA-AAAGTTCTATAATTGAAT-CTTTTGGAAA-------------------------------------------------------------------------------------------------------------------------------------------TTTTAGAATCTCATCTATA-T--ATGTCCATCTTA---ATCTCT---------T-------CCAGACAATAGCGGTATAGCGGA-----GGATCAGACACGGAAGCGGGAGATTCGATTG | |
| droKik1 | scf7180000302696:1337717-1338210 + | GATGCAGTTGG------ACCCGGCGTACTAC---CTATCCAATCGGATATCTTACAACACCAGTAATA--GTCCCA-TCCAAA----------------GA---ACTTATAT------------CTATAGACCTTAAATAGC--------GCTAGAATTGAACT--CCTGCA------------------------------------------------------------------------------ACTCGAACTTGCTGATACAATCGATTTAAATAGTATTATATTTGTACATCAAATTATGGTTCTTTTCGTAATCCCAAATATGTATAATTAAATTTATTTCAAAGTTAAACGTAAAATTATTAAAATTAAACTTTTCATGTAACTATTTTGTCTATTGCTTTTATAAAAAGAATGAATCTTTTTGAATTCCAAGCTAT---------------------CCAAGTTCGAGT---------------------AGA---------------------------------------------------------------------------------------------------GTTAA---------------------------------------------------------------------------------------------TTCGA-ATCCCTTTCGAGTTACAG-AGAGTTCGACTTTTGGGGA---------------------------------------------AATTAAAGCACACACAA----TCG---GTT----------------------------AA--------------------------------TCGGATAACTAATC-----------------ATCTGTTC---TTATTCCTATTTCT---------T-------GCAGACAATAGCGGGATAGCGGA-----GGATCAGACGCGCAAACGCGAGATCCGGCTG | |
| droFic1 | scf7180000453829:1361922-1362070 - | TAAATAACA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACAGTTCGACTTACAAAGGGGTTTCGCCTTGA-AAAG-A------------------------------------------TAG----GC---------------------------------TT-------TAGAATTTGG------------------------------------TAACTAAT-----------------CATATTT--CCACCATAT---TCTAT---------T-------GCAGACAATAGCGGTATTGCGGA-----GGATCAGACCCGTAAGCGCGAAATCCGGCTG | |
| droEle1 | scf7180000491006:509851-509924 + | GATGCAGTTGG------ATCCCACGTACTAC---CTGACCAATCGGATGTCCTACAACACCAGTAATATAGCCTCGGTCCAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A----------------------- | |
| droRho1 | scf7180000778010:45602-45703 + | ACTTAAAAAAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTGAAATTTAAAAA---TCATA-TTTTATGTTTAAATGCATGGG-------------------------------------------------------------------------------------------------------------------------------------------T---------------------------------TTGTGAAGCTACAATTAAA------------------------------------TATATAAA-----------------TATATTT--ATA-----------------------------------------------------------------------------ACAAGTGCA-ATT | |
| droBia1 | scf7180000302432:988468-988555 - | GATGCAGTTGG------ACACCTCGTACTAC---CTGTCCAATCGGATGTCCTACAACACCAGTAATA--GCCTCGCTCCAGC----------------AGCGAGTCGAAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTG | |
| droTak1 | scf7180000415395:275269-275432 + | AAAATTTCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTCAGTTCGAGTTACAG-GCAGTTCCAATCTTTGATA---------------------------------------------GGT----TAGCGCTTTAGCGACCG-----------------------------TTTTGTG--------------------------------CTAATTATCTA------C---------TTTG-T--TTTT---CTTAA---ATCTCT---------T-------GTAGACAACAGCGGGATAGCGGA-----AGATCAGACCCGCAAGCGAGAGATCCGGCTG | |
| droEug1 | scf7180000409528:451464-451862 + | GATGCCAATGG------ATCCCACGTACTAC---CTGTCCAATCGGATGCCCTACAACACCAGTAATA--GCCCCGGTCCAAT----------------CA---AC----TT--------------------CTTGAATTACATATAGGAGTTGG--TGGAACTTTTCTGTAAATCGATTTTTTCTTAAGTTGGTTGTAGACGATGATCCAG----------------------------------------------TTGCTCATGCGGTTGAAT-----------------GAACGTCT-------------------------------------------------------------------------------------------------------------------------------------AATCTCAGGCTAAACTTAAGTATTAACGTAAATCTCAAGATCAAGAAAAACTTCATAAAGATCAAATAAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTCAGATCGATTTACAG-GGAGTTCCACTTTAA-AGAA---------------------------------------------AT----CTACAATCTA----AAG---ACT----------------------ACAATTTA--------------------------------CTGATCAAT---------------------C-T--ATCT---TTTTGCATATATTT---------T-------ACAGACAATAGCGGGATAGCGGA-----GGATCAGACCCGTAAGCGAGAGATCCGGCTG | |
| dm3 | chrX:18269367-18269439 + | GATGCAATTGG------ATCCCACGTACTAC---CTGTCCAATCGGATGTCCTACAACACCAGTAATA--GCCTCGCTCCAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA | |
| droSim2 | x:17367441-17367581 + | GTTCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTTACAG-AGAGTTCCGCTTTGGGAGA---------------------------------------------TGT----GCACACATTG----CTG---ACT----------------------------GA--------------------------------CTGATCATCCAATCCTTGAATCCTTTT--------------------TTC---ATT---------T-------GCAGACAACAGCGGGATAGCGGA-----GGATCAGACCCGCAAGCGCGAGATCCGGCTG | |
| droSec2 | scaffold_8:579264-579404 + | GTTCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTTACAG-AGAGTTCCGCTTTGGGAGA---------------------------------------------CGT----GCACACATTG----CTG---ACT----------------------------GA--------------------------------CTGATCATCCAATCCTTGAATCCTTTT--------------------TTC---ATT---------T-------GCAGACAACAGCGGGATAGCGGA-----GGATCAAACCCGTAAGCGCGAGATCCGGCTG | |
| droYak3 | 3R:25686943-25687109 + | ACAAAGTTTAG------TT-GAAAGAATGAA---ATAATAAATTCA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTATTAGAATAGAAGTAACAAATTGTTTAAAATTAGTGACAATTTGTAAAAGTAACATTTCAAATTTATCGAAAGCCT-------------------------------------------------------------------------------------------------------GTGTGCAAATTTTAA-GCACTTTCATTTTAAAAT-CAT---------------------------------------------TTC----A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATTTAAAATA | |
| droEre2 | scaffold_4690:8590622-8590689 + | TGTATCCTTGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT---------T-------ACAGACAACAGCGGCATAGCGGA-----GGACCAGACCCGTAAGCGCGAAATCCGGCTG | |
| dp5 | XR_group8:7996351-7996426 - | TTTAAATAATT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-GTATATTTAAAAAAATTTAACTATTTTTTTTTTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATAAAATAAATAAAAATATATATAAAAAA | |
| droPer2 | scaffold_39:506698-506758 - | TCTGTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-------ACAGACAACAGCGGCATAGCGGA-----GGATCAGACGCGCAAGCGGGAGATCAGACTG | |
| droWil2 | scf2_1100000004515:1511886-1512273 + | CCTGCATATGGGTCCAGAATCGGCCTACTAT---ATATCCAATCGGATAGCCTACCAAACCAGTAATA--TCCATAC-CAAT-----------------------TCTAAGT------------CTAGGGA--------------------------------------------------------------------CACAATGAGCCAGCCTGAAGTTCGGAAATA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAAAAAGG--GCGCATTAGTTTTT--------GAAAATTATATGCAAATGACAAATGATTTG--------------------------------------------------------------------------------------------------------------------------------------------------------------GAGTTATAG-TAT---CGGCCGATCAAT-ATTTTCCGAACTTCAGGATCTCATTGTTTTGATTCGAAAGAGAAATGT----GATAAC------AATTGTCTTCTAACTATGAAAATCTATCAATCTTGCTATAAATATATGTG-----------------------TA-TATAACCAATT-----------------ACCTCTGT---TCGTTC---TTTAT---------T-------ATAGATAACAGCGGGATTGCAGA-----GGATCAGTCGCGCAAGCGGGAGATACGTTTG | |
| droVir3 | scaffold_12928:4487326-4487478 - | TCTAAAAATAA------CTAAAGCAA----------------------------------------------------------------------------------AAAATTAAAAAGAAA-----------------------------------------------------------------------------------------------------ACAAATTACTTCAAATTTAAAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAATTTTAAA---------------------------------------------------------------------CTTTAAGCAAAGCCAAAGAACGGTTTAAGACAAAAAACAAAAACA-AA-----------------TGAACTTAAGCTA-----AATAAAATA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTATTT---TTTTT---------T-------G-------------------------------------------------------CGA | |
| droMoj3 | scaffold_6473:3890381-3890655 + | dmo_666 | GCTGCACATGGCGTCGGACCCGGGCTACTTT---CTATCCAGTCGGATAGCATACAATACCAGTAAAT--GCCACAAAATACA----------------A---------------AAAAAAAA-----------------------------------------------------------------------------------------------------AAAAAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATAAAATAAAATAAA-----------------------------------------------------------------AAATAAAAAAAGAAACAAACAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA---------------------------------TT-------TATAACTGAG--------CCAAATACTTATACTCTTCTTTCTTCTTCTTCTTCTCTCT----CTCTCT--------------------TCC---ACTCTCTGTGTCGGCACCTCTTAGATAACAGTGGCATTCCGGAGGAGAGGATCAGACACGCAAGCGAGAGATTCGACTG |
| droGri2 | scaffold_15081:798026-798304 - | GCTGCACATGGCGCCAGACCCGGCCTTCTATTTGCCATCCAATCGGATAGCATACAATACCAGTAAAT--GCCAAAAAC-A-TCAGCTACAAATAGCAAAA---AAAAAAAAAAAGAAAGAAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAAAATAAAATCAAAATCAAAATAAT-------AATAATAATACACAA-----------------------------------------------------------------------------------------AAATAAAA-----------------TGAATAG-------------------------------------------------------------------------------------------------------------------------------------AA---------------------------------AT-------GAAAACTAA--------------------------------TTCTTGTTCTTCTCTTGCAACGCATTC--------------------TAT---TTG---------TGCA--ACACAGATAACAGCGGCATTGCGGA-----GGATCAGACGCTCAAGCGGCAAATACGATTG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 02:09 AM