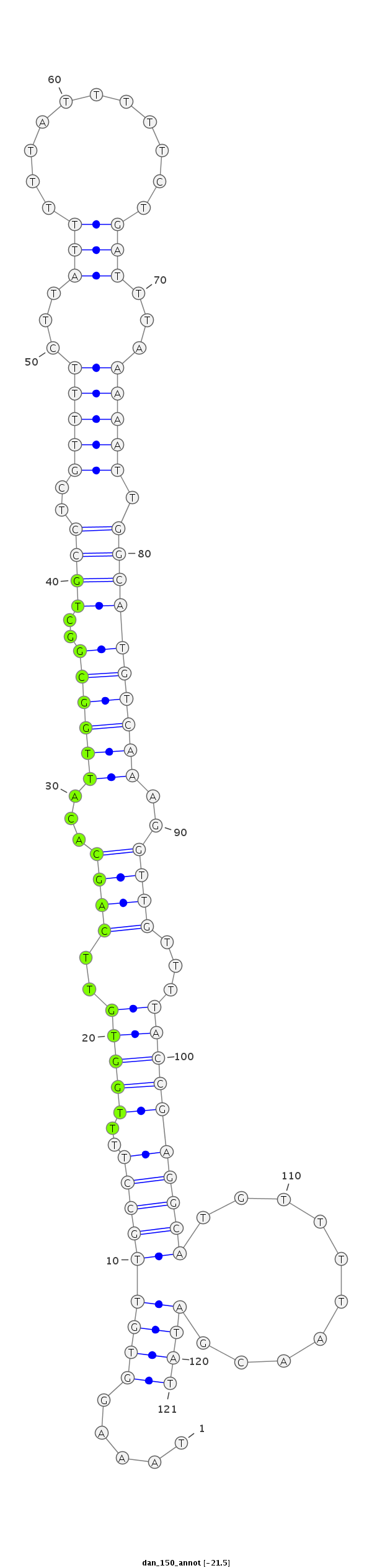
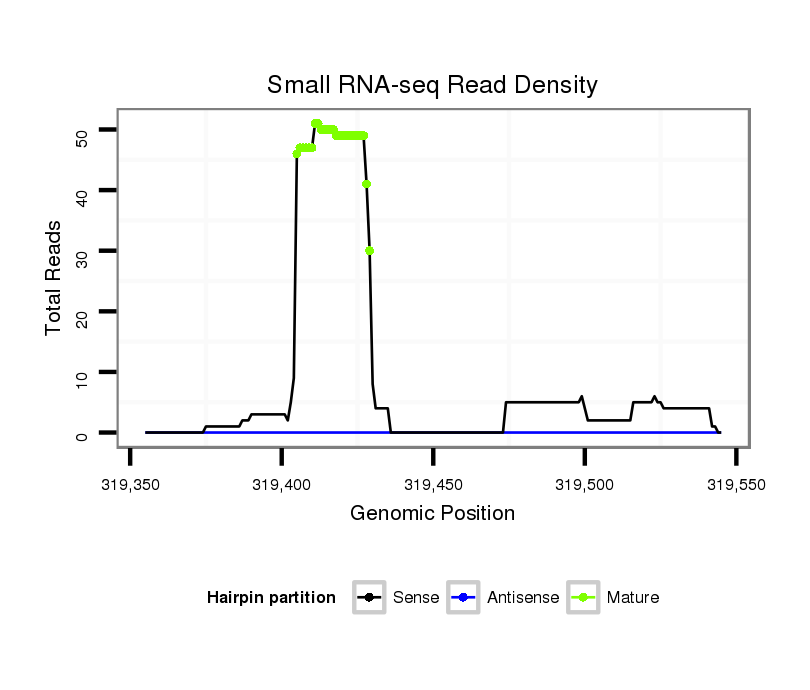
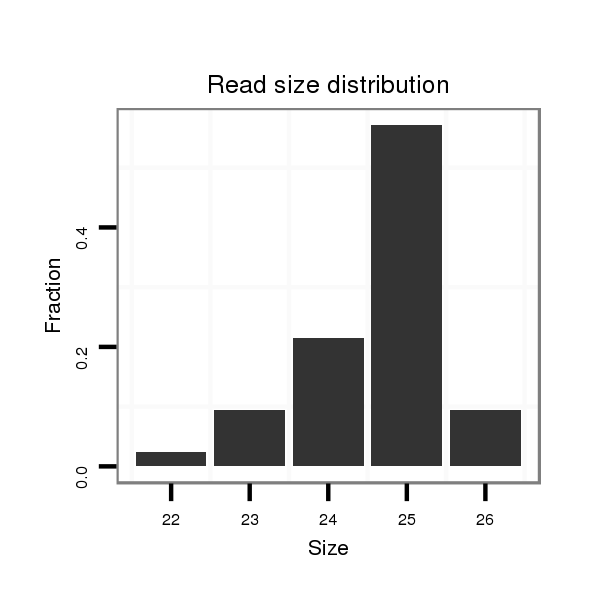
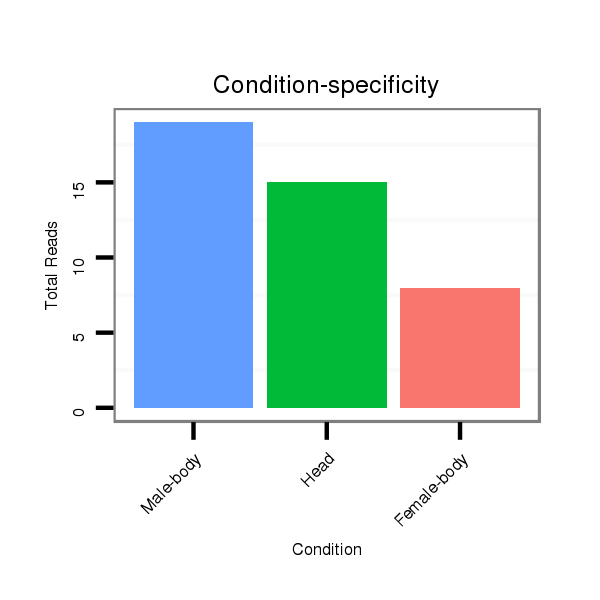
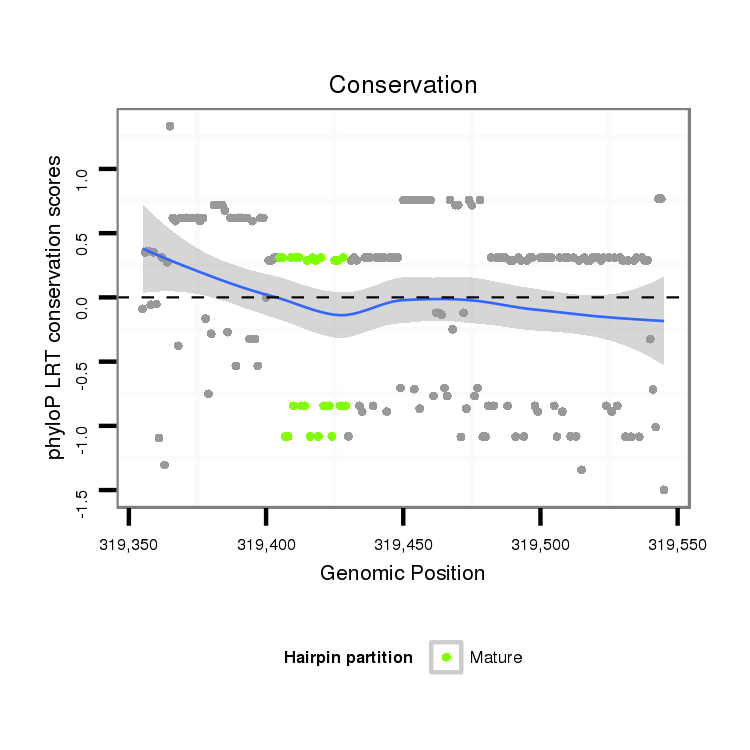
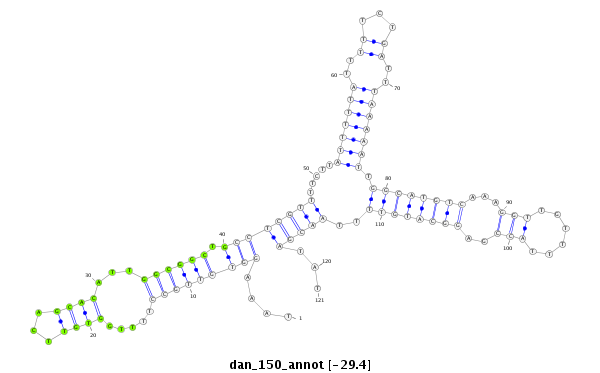
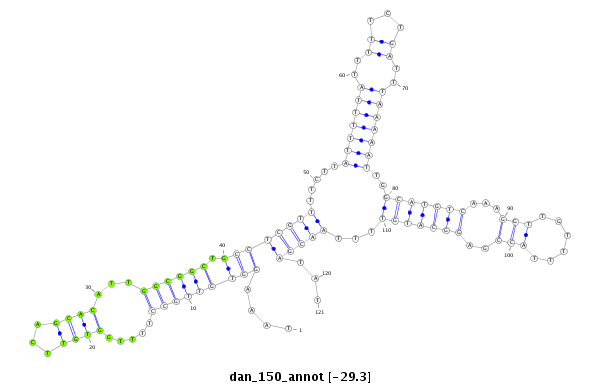
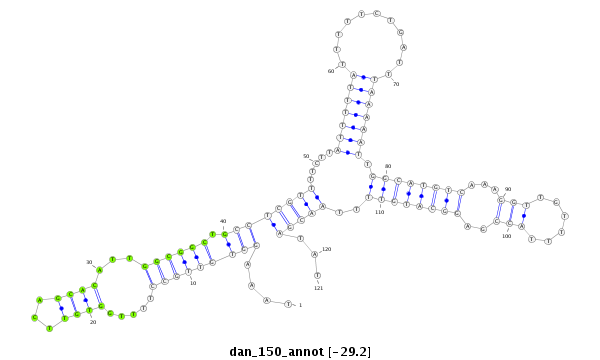
| ..................................................TTGGTGTTCAGCACATTGGCGGCTG.................................................................................................................... |
25 |
0 |
1 |
20.00 |
20 |
5 |
12 |
3 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGCT..................................................................................................................... |
24 |
0 |
1 |
9.00 |
9 |
7 |
2 |
0 |
0 |
0 |
| ........................................................TTCAGCACATTGGCGGCTGCCTCGT.............................................................................................................. |
25 |
0 |
1 |
4.00 |
4 |
3 |
0 |
1 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGCTGC................................................................................................................... |
26 |
0 |
1 |
4.00 |
4 |
1 |
0 |
3 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGC...................................................................................................................... |
23 |
0 |
1 |
4.00 |
4 |
3 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGCTGT................................................................................................................... |
26 |
1 |
1 |
4.00 |
4 |
3 |
1 |
0 |
0 |
0 |
| .................................................................................................................................................................TGGTTTCTTATTGTGGGTCATTGGTG.... |
26 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGCTGTT.................................................................................................................. |
27 |
2 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
| .......................................................................................................................TCAAAGGTTGTTTTACCGAGGCATGT.............................................. |
26 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ................................................TTTTGGTGTTCAGCACATTGGCGGC...................................................................................................................... |
25 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| .......................................................................................................................TCAAAGGTTGTTTTACCGAGGCATGTT............................................. |
27 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| .................................................TTTGGTGTTCAGCACATTGGCGGCTG.................................................................................................................... |
26 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
| ...................................................................................................................................................TAACGATATCACCGTGGTTTCT...................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGATGT................................................................................................................... |
26 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGA...................................................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ...................................................TGGTGTTCAGCACATTGGCGGC...................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ................................TTTTAAAGGTGTTGCCTTTTGGTGTT..................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGCTCC................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................TTATTGTGGGTCATTGGTGTA.. |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCTGCTG.................................................................................................................... |
25 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................TTTGGTGTTCAGCACATTGGCGGCT..................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ....................TGTGTCTTTTCTTTTTAAAGGTGTTGC................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................................................TCAAAGGTTGTCTTACCGAGGCATGTTT............................................ |
28 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCGGCACATTGGCGGCTG.................................................................................................................... |
25 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................TTAAAGGTGTTGCCTTTTGGTGTTCAGT................................................................................................................................. |
28 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................TAAAGGTGTTGCCTTTTGGTGTTCAGCA................................................................................................................................ |
28 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................................................................................................................TTATTGTGGGTCATTGGTGTT.. |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................TTTTAACGATATCACCGTGGTTTCTTA.................... |
27 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ................................................TTTTGGTGTTCAGCACATTGGCGGCT..................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................................................TCAAAGGTTGTTTTACCGAGGCATGTTT............................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TTTGTGTTCAGCACATTGGCGGCT..................................................................................................................... |
24 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCCCATTGGCGGCTGC................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGCTT.................................................................................................................... |
25 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................................................................................................TTATTGTGTGTCATTGGTGTA.. |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................TTTGGTGTTCAGCACATTGGCGGC...................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................TTGGTGTTCAGCACATTGGCGGCTGCT.................................................................................................................. |
27 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .........................................................................................................................................................TATCACAGTGGCTTCTTTTT.................. |
20 |
3 |
19 |
0.32 |
6 |
0 |
0 |
0 |
6 |
0 |
| .................................................................................................................................................TTTCAGGATATCAACGTGGT.......................... |
20 |
3 |
8 |
0.13 |
1 |
0 |
0 |
0 |
0 |
1 |
| ......................................................................................................................................CGGAGGAATGGTTTAACG....................................... |
18 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |
| .........................................................................................................................................................TATCACAGTGGCTTCTTTT................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
| .........................................................................................................................................AGGCATGGATTCACGATAT................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |