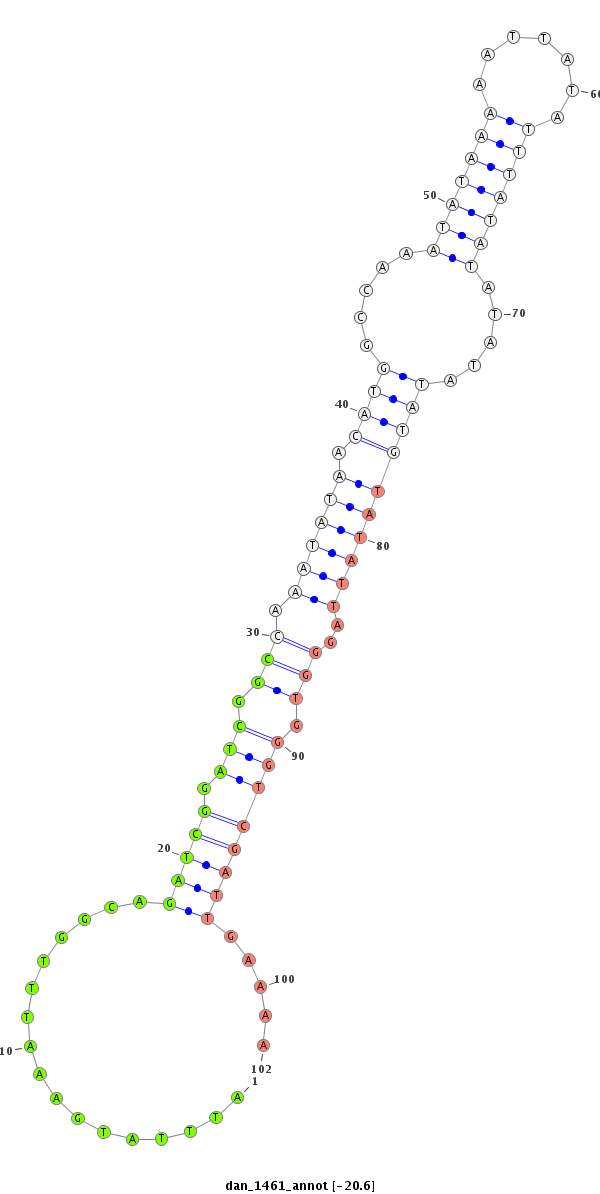
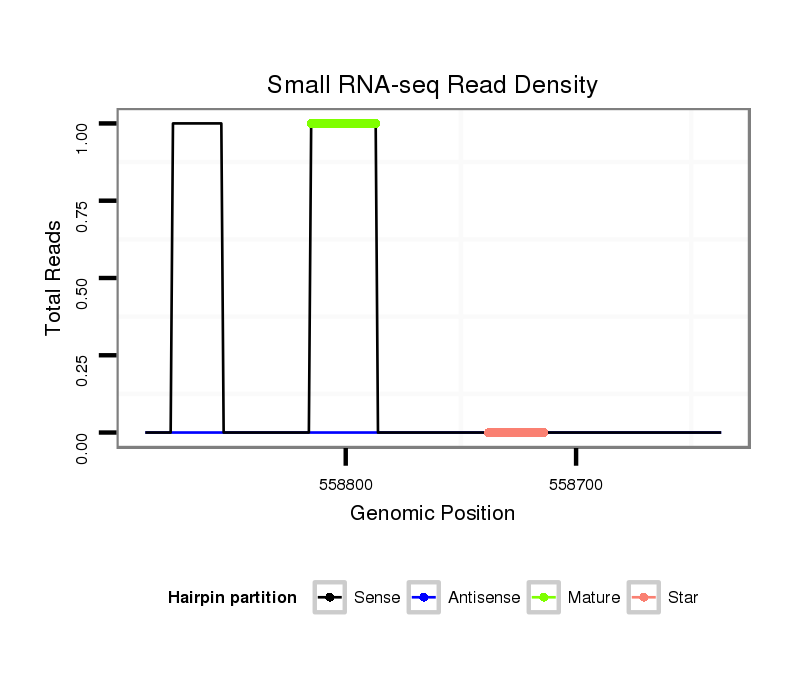
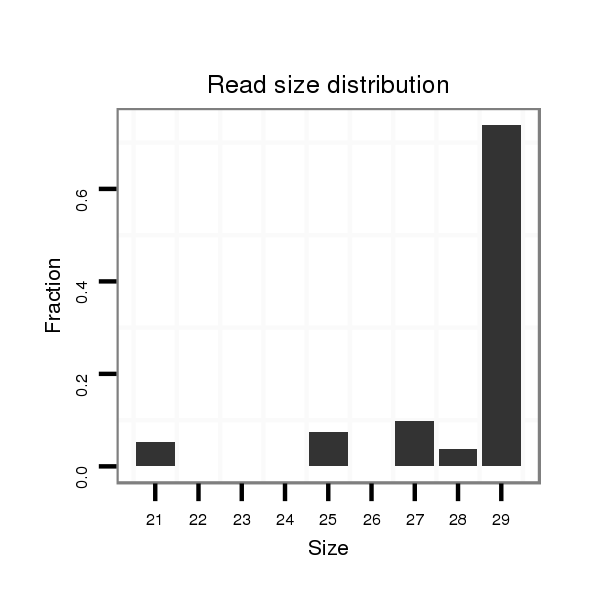

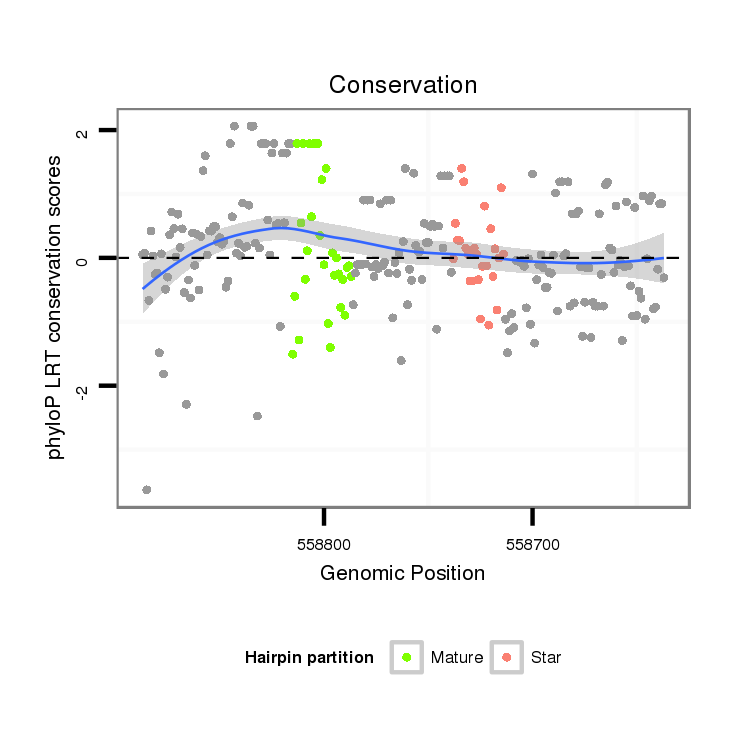
ID:dan_1461 |
Coordinate:scaffold_13230:558687-558837 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.6 | -20.6 | -20.6 |
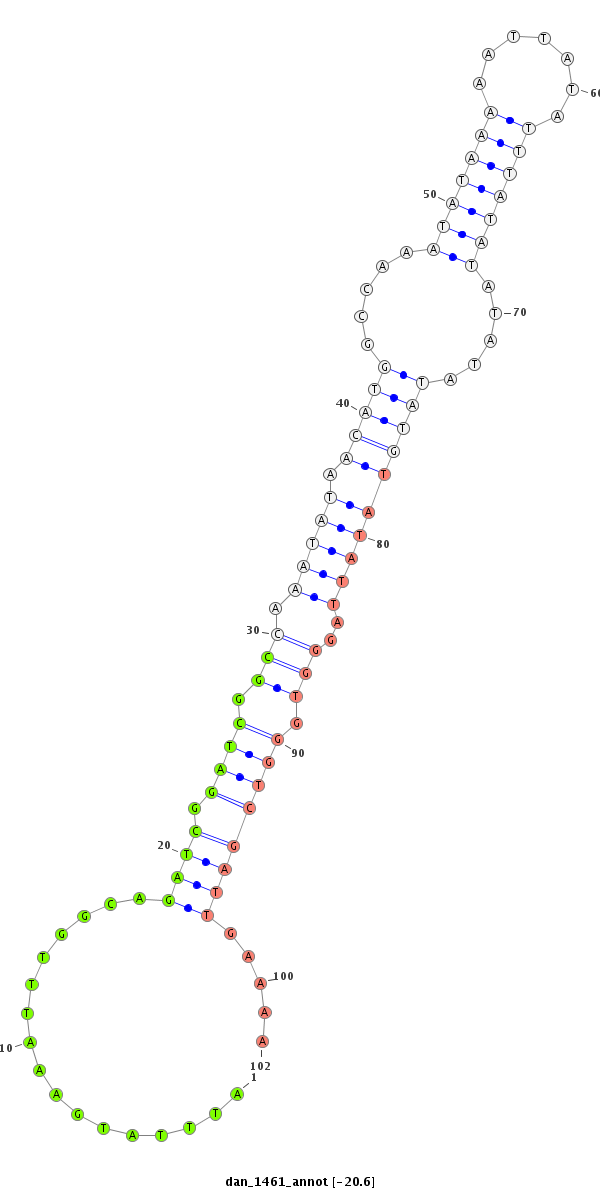 |
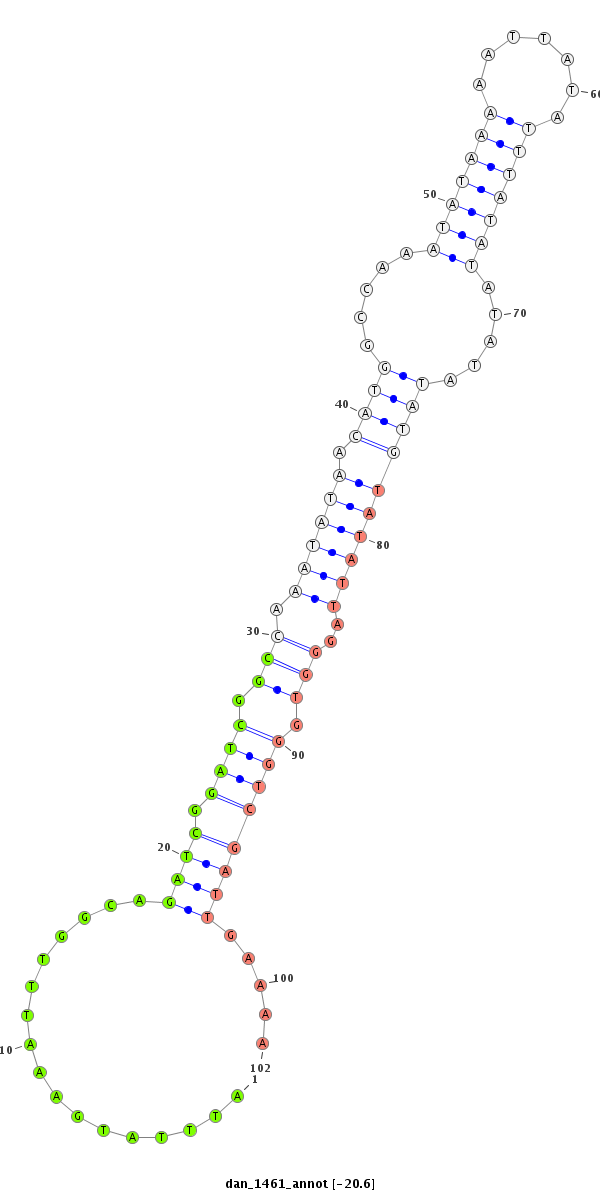 |
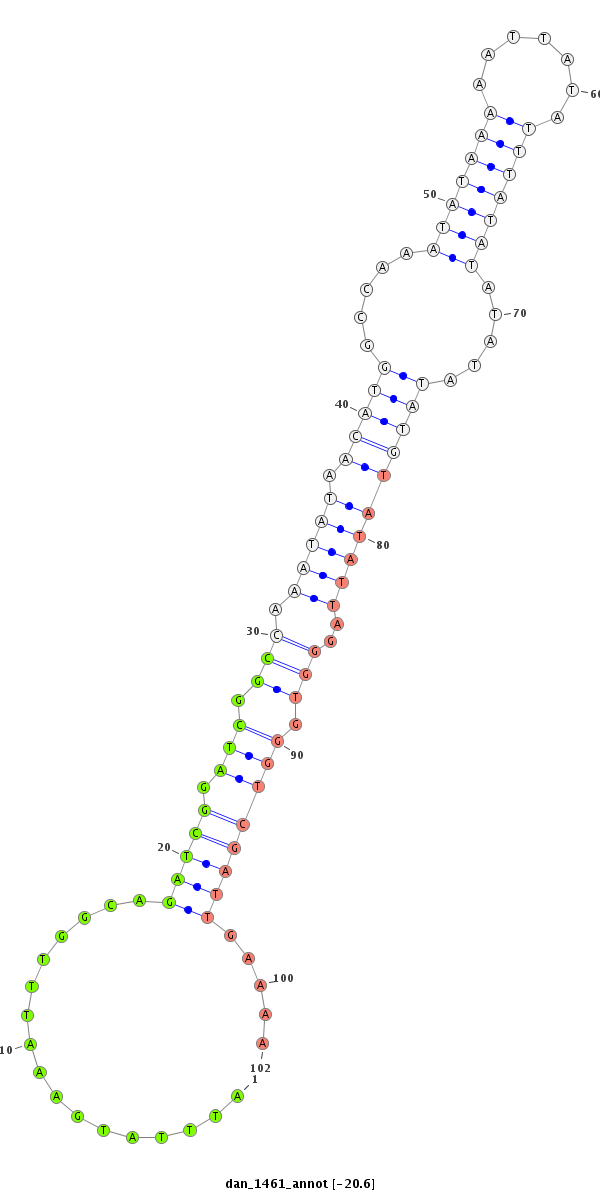 |
CDS [Dana\GF15848-cds]; exon [dana_GLEANR_1661:1]; intron [Dana\GF15848-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| Hoana1 | DNA | hAT-hobo | - |
| -------------------------------------------#######--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTTAAAAATAGCAAAGTGGTGCCATATTCGATCGTTCAGTTATATGGCAGGTATGAAATACGGTCGGCCGATATTTATGAAATTTGGCAGATCGGATCGGCCAAATATAACATGGCCAAATATAAAAATTATATTTATATATATATATGTATATTAGGGTGGGTCGATTGAAAAATCGCTTAGGCACATATGATTTTCGGATTCTAGGGATCAAAATAAGAAACTTTGCTCAAGAGACCATACCTCTAAAA ************************************************************************.................(((((.(((.(((.((((((.((((.....(((((((.......))))))).....))))))))))..))).)))))))).....***************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V055 head |
V039 embryo |
V105 male body |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................ATTTATGAAATTTGGCAGATCGGATCGGC...................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............AAAGTGGTGCCATATTCGATCG......................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGCTCAAGAGACCATACCTCT.... | 21 | 0 | 20 | 0.50 | 10 | 10 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TTGGCAGATCGGGACGGC...................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GCACATATGATTTTCGGATTCTAGG........................................... | 25 | 0 | 20 | 0.45 | 9 | 7 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GCACATATGATTTTCGGATTCTAGGGA......................................... | 27 | 0 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GCACATATGATTTTCGGATTCTAGGGAT........................................ | 28 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................GATATTTAAGAAATTTGGCGGG................................................................................................................................................................ | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TTTGCTCAAGAGACCATACCT...... | 21 | 0 | 20 | 0.15 | 3 | 2 | 0 | 0 | 0 | 0 | 1 |
| ................................................AGGTGTGCAATACGGTCGC........................................................................................................................................................................................ | 19 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................GTTTGAAAAATGGCTTCGGC.................................................................. | 20 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................TATATTAGGGTGGGTCGATTGAAAA............................................................................. | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................TGTATATTAGGGTGGGTCGATCGAAAA............................................................................. | 27 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................GTATATTAGGGTGGGTCGATTGAAAAA............................................................................ | 27 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............AGGGGTGCCATATTGTATC.......................................................................................................................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................TGTATATTAGGGTGGGTCGAT................................................................................... | 21 | 0 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................TGGCAGGGTTGAAATTCGG............................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GCACATATGATTTTCGGATTCTAGGG.......................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TAAGAAACTTTGCTCAGGAGACCATA......... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GCACATATGATTTTCTGATTCTAGGG.......................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGCTCAAGAGACCATACCTCTAA.. | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................ACTTTGCTCAAGAGACCATA......... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CTTGAAAGATCGCTTCGGC.................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................TATATTAGGGTGGGTCGATTGAAAAAT........................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................GGCACATATGATTTTCGGATT................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................TATATTAGGGTGGGTCGATTGAAAAATC.......................................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................TCGTTCAGTTATATGGCAGGT....................................................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GCACATATGATTTTCGGATTCTAG............................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................GAAACTTTGCTCAAGAGACCATACCTC..... | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TAAGAAACTTTGCTCAAGAGACCATA......... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................TAGGGATCAAAATAAGAAACT.......................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TTTGCTCAAGAGGCCATACCTC..... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AAATTTTTATCGTTTCACCACGGTATAAGCTAGCAAGTCAATATACCGTCCATACTTTATGCCAGCCGGCTATAAATACTTTAAACCGTCTAGCCTAGCCGGTTTATATTGTACCGGTTTATATTTTTAATATAAATATATATATATACATATAATCCCACCCAGCTAACTTTTTAGCGAATCCGTGTATACTAAAAGCCTAAGATCCCTAGTTTTATTCTTTGAAACGAGTTCTCTGGTATGGAGATTTT
*****************************************************************************.................(((((.(((.(((.((((((.((((.....(((((((.......))))))).....))))))))))..))).)))))))).....************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V105 male body |
V039 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................CGGTATAAGCTAGCAAGTCAA.................................................................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................AACGAGTTCTCTGGTATGGAG..... | 21 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 |
| ...............................AGCAAGTCAATATACCGTCCA....................................................................................................................................................................................................... | 21 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................AGCGAATCCGTGTATACTAAAAGCCT.................................................. | 26 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TTGAAACGAGTTCTCTGGTAT......... | 21 | 0 | 20 | 0.10 | 2 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................ACGAGTTCTCTGGTATGGAG..... | 20 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................ATCCGTGTATACTAAAAGCCT.................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................TTAGCGAATCCGTGTATACTAA........................................................ | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TGAAACGAGTTCCCTGGTATG........ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CCCTAGTTTTATTCTTTGAA......................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................AAAAGCCTAAGATCCCTAGTTTT................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................CGAGTTCTCTGGTATGGAGAT... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CTAACTTTTTAGCGAATCCGTGTATACT.......................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................AGCCTAAGATCCCTAGTTTTAT................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................TGCGAATCCGTGTATAGTAAAAGCC................................................... | 25 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CCCTAGTTTTATTCTTTGAAA........................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................CTAGCAAGTCAATATACC............................................................................................................................................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................GAGTTCTCTGGTATGGAGA.... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13230:558637-558887 - | dan_1461 | TTTAAAAATAGCAAAGTGGTG-----------CCAT----------------------A--TTCGATCGTTCAGTTATATGGCAGGTATGAAATACGGTCGGCCGATATTTATGAAATTTGGCAGATCGGATCG----------------GCCAAATATAACATGGCCAAATATAAAAATTATATTT----A----------------T----------ATATATATATGT----------ATAT-----------------TA-GGGTG--------GGTCGATTGAAAAATCGCTTAGGCA---------CAT------ATGATTTTCGGATTCT--------------------------------------------------------AG-GGATCAAAATAAGAAACTTTGCTCAAGAGACCATACCTCTAAAA |
| droBip1 | scf7180000392463:2439-2685 + | TTCAAAATCACGAAAGTTGGG-----------TCAT----------------------T--TCCGATCGTTCAGTTATATGGCAGCTATAGGATATAGTCGGCCGATCCTTATGTAATTTGGCATGTCGTAATGTTTTGCCAAA---------------------------AATATAA-----TT----TTGGACAAAATTATAATTTTGGTCAAAAGTGTGCT------ATAGCTGCCA-------------------------------TATAACTGAACGATCGGAAATGGTTTTTGGTA---------GAA------AT-----------------------------------------------------ACCAACTTTCGTATTTTTGAAGATAGAAGCTTGATACTTTGTTTGAG----------------- | |
| droKik1 | scf7180000302355:1053216-1053404 - | TTGAAAAACAACGAAGTTATA-----------ATGT----------TTTTTCTATTAATTTCCCGATCGTT----CCTATGGCAGCTATAAGATATAGTCGTCCGATATTCATAAAATTTT----------------TACCAAAATTCTGAAATAATATAATATGGTCATATGT---------------------------------------------AAAAA------ATAGTGCACA-------------------------------TAT---------GTTGAAAAACAGCTAAGTT-----------------AT-----------------AATT-------------------------------------------------------------------------------------TTTTTCTAAAAAT | |
| droFic1 | scf7180000454095:19058-19123 - | TATGACGATTTCCAT--------------------------------------------------ATTGTT----CATATGACAGCTATATGATATAGTCGTCCGATTTTGATGAAATTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000489694:22705-22833 - | TTAAAAAACACCAAAGTTATG-----------TCAA----------------------T--TCCGAACGTTTAGTTATTTGGCAGCTATACGATATAGTCGGCTGATCCTTATGTATTTTG-TAGTTCGGTTCAATTGACAAAA---------------------------AATACAATCCATTCTTATTT--------------------------------------------------A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droRho1 | scf7180000775924:3736-3980 + | TTTAAAAACACCAAAGTTATG-----------GCAT----------------------T--TCCGATCAATCAGTTATATGGCGGCTATAGGATATAGTCGGCCGATCCTTATGAAATTTGGTAGGTTGGATTGATTGGCCAAA---------------------------AACAAAACCCATAGTTA--------------------------------------------------GGAATTTGGTATTTTTTTGAGTATTAAGAATG--------CATCATATG--------------------------------------------AATAATTTTGGAGAATATAATTTACAAAATATTCTGCAAGGGTATATCAACTTCGG-------------------------CTCCGCCCAAAGTAGCTTTCCT------ | |
| droBia1 | scf7180000302506:53081-53185 + | TTAGAAAACACCGAAGATTTA-----------ATTTTATCATATTATTTCACCACTAAATTTTTGATCGTT----CCTATGGCAGCTATATGATATAGTCGCCCGATTTTGATAAAATTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000413235:29467-29595 + | TACATACATT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAT----A----------------C----------ATACATGTATAT----------ACAT-----------------TA-GGGTG--------GGTCGATTTGGAGGTCAACAAATCG---------TAG------GGGGGGAACGTATTCT--------------------------------------------------------AG-AGACTATTCTAAGCAACATTGCCAAAGAAACTATGCCTTGGAAA | |
| droEug1 | scf7180000407619:547-622 - | ATATAATATATATA-GATAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATA------TATATATATA-------------------------------TAA---------GTAGAGAAACCG-----------------TATATACAT-----------------AAAATACATAAAAT---------TTT---------------------------------------------------C------------------------ | |
| dp5 | 2:25264180-25264290 + | TTTGAAGGAT-------------------------------------------------------------CAGAACTATCGTATCTATTTGATATGGTTGACCGATTCTACTGAAATTTGGTACAGTCACCCG----------------GCTATTAACAATCATGCTAAACTACAAATTTAAAT----TTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_56:95400-95435 - | ATATATATATATAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAT----A----------------T----------ATATATATATGT----------ATAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000000514:5726-5866 - | TTTAAAAATAACAAAGTTATTGACAAAAGTCACTCT----------------------T--TTCGACCGTCCGTTCCTATGGGAGCTATATGATATAGTCAGCCGATCTTGATCAAATTTGGCATAGTCGTTTATATGTAAAAT---------------------------AAACTCATCAATATTAAATT--------------------------------------------------T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droMoj3 | scaffold_6500:21953045-21953188 + | CCTATTATAATAATACCTA------------------------------------------------------------TTATAATAATAAT-------------------------------------------------------------------------------------------ATAT----A----------------T----------ATATATATATATATATATATGTATATTGTTTTTTTTTCAGCATTTTTAAATTTGA---------TT-----------------GCAGAAGGTGTAT------TTAATTTATGAATTCTTAAGATGAATAAAAT-----------------------------------------------------------ACTTT------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/18/2015 at 01:58 AM