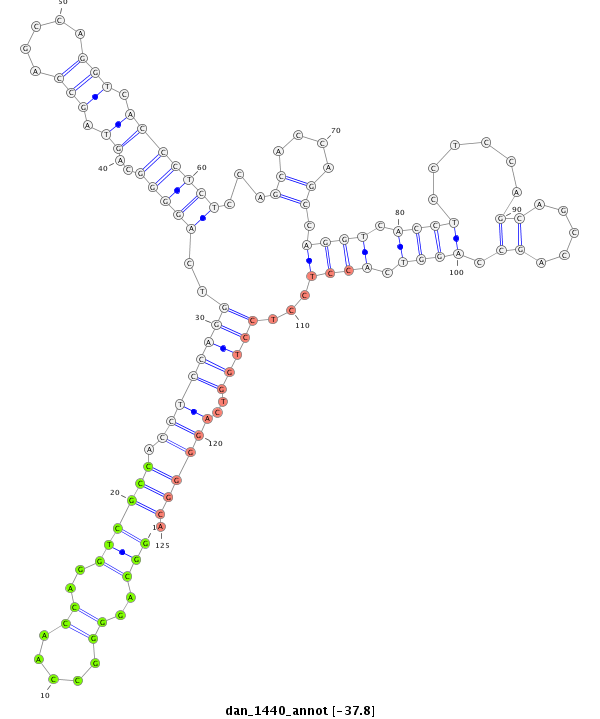
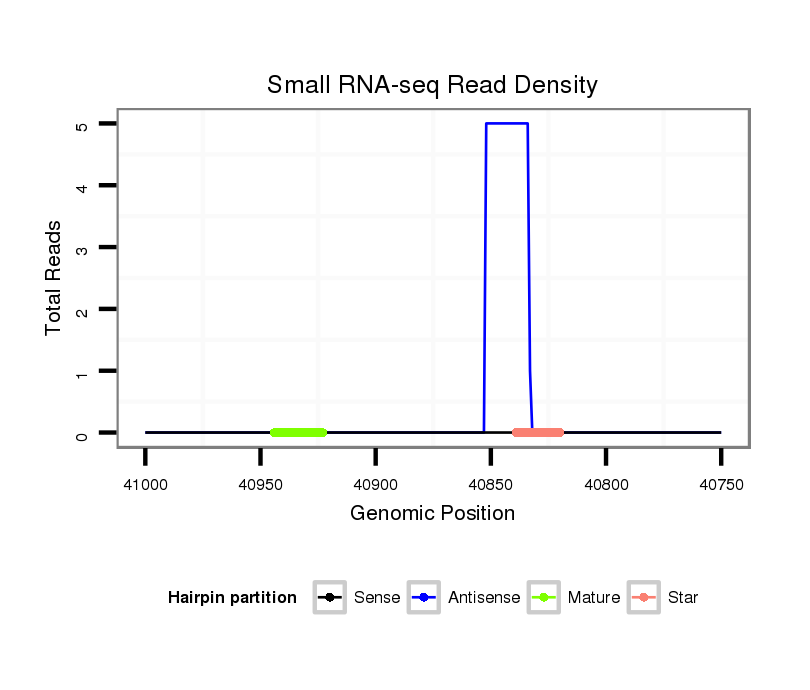
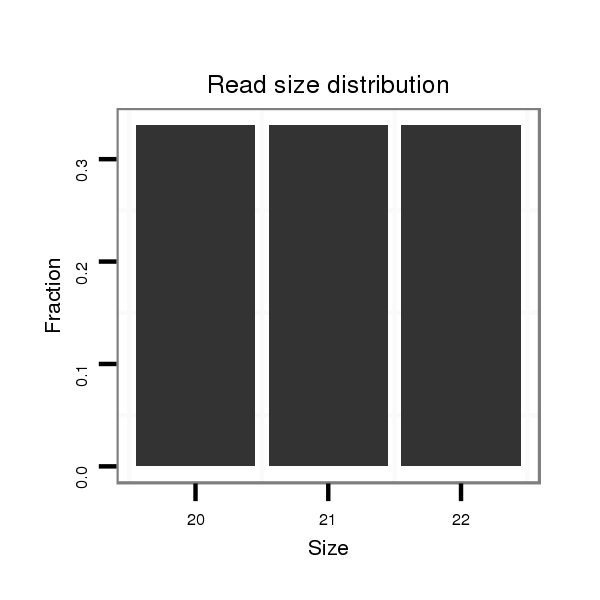
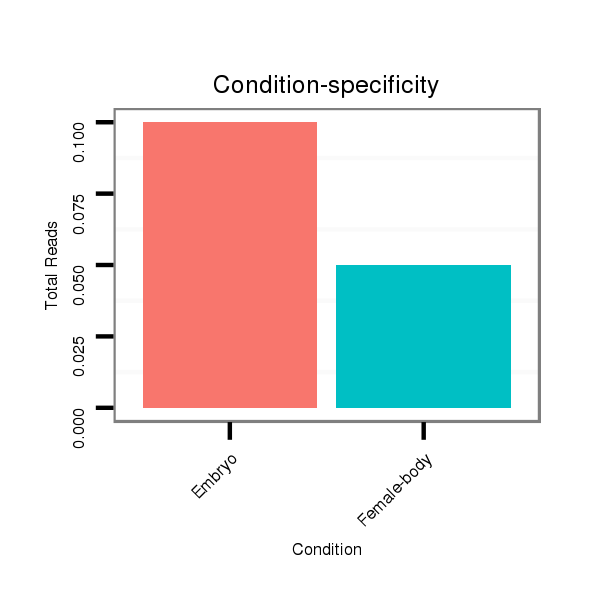
ID:dan_1440 |
Coordinate:scaffold_13150:40800-40950 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -37.6 | -37.6 | -37.5 |
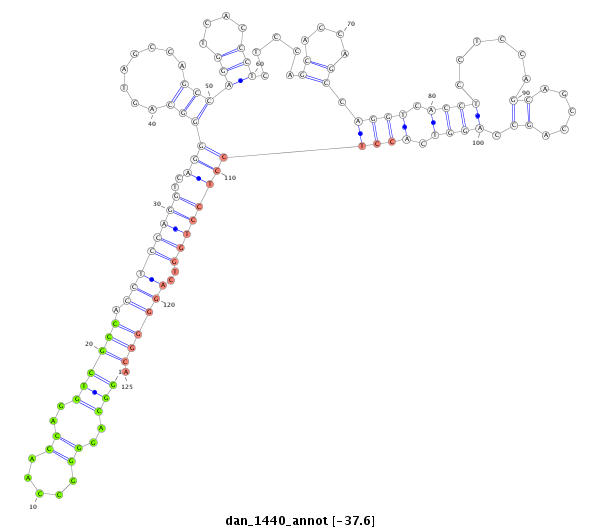 |
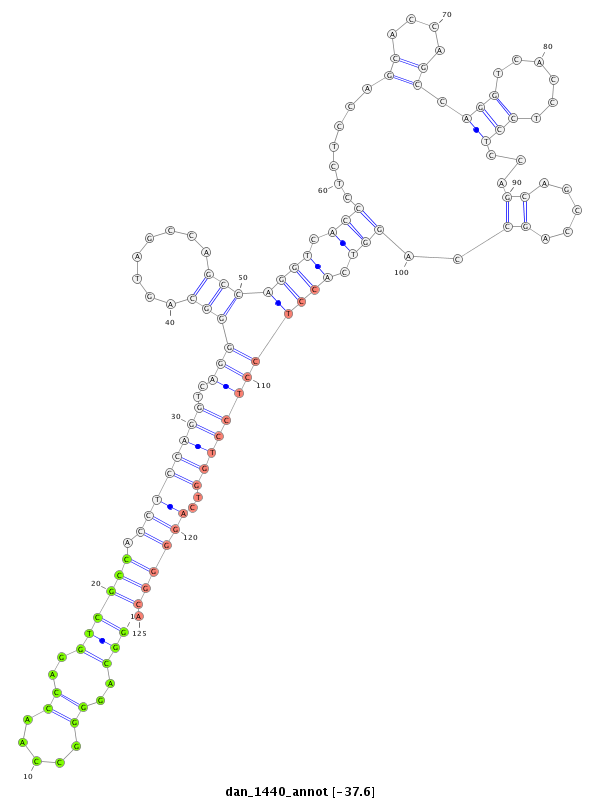 |
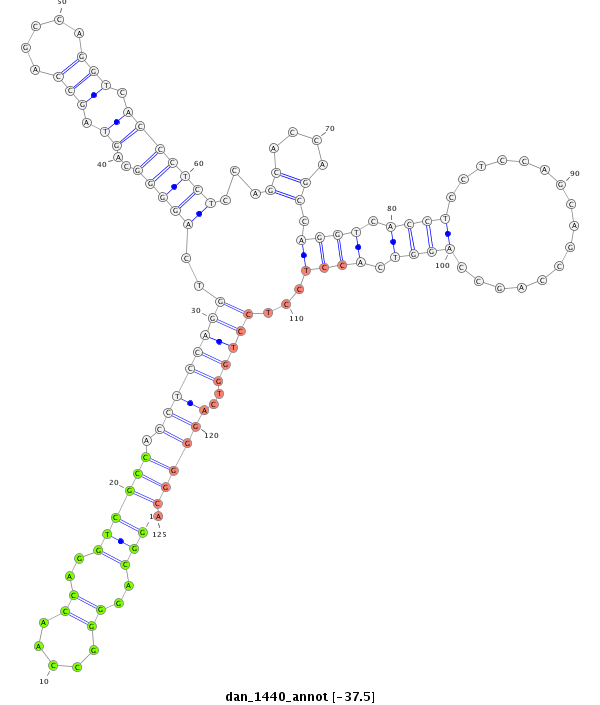 |
exon [dana_GLEANR_20368:3]; CDS [Dana\GF18978-cds]; intron [Dana\GF18978-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CACCCAGCCAGGTCACCGTCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGTGGCAGGGGCCAACCAGGTCGCCACCTCCAGGTCAGGGGCAGTAGCCAGCCAGGTCACCCTCTCCAGCACCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGGTCACCTCCTCCTGGTCAGGGGCAGGGGAGTGCACCTCCTCCAGTCGCCGGCCATTCCGGCCCGTCTCCCCGCCGAGGGGAAAGTGCCTGAGGG ********************************************************(((..((.....))..)))(((.((((((((..(((((..((.(((.....))).)))))))...((....)).((((.((((......((.....)).)))).))))...)))))..)))))).********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V055 head |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................CTCCTCCAGTCGCCGGTCATT...................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ....................................................................................................................CGACAGCACCACCCAGGTCACCT................................................................................................................ | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ...........GTCTCCGTCTCCAGCAGCCAGC.......................................................................................................................................................................................................................... | 22 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................................................................ATTCAGGGGCAGGAGAGTGCA............................................................ | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................................CGAGGGGAAAGTGCCTGAGG. | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ........................................................GGCAGGGGCCAACCAGGTCGCC............................................................................................................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ................................................................................CTCCAGGTCGGGCGCAGT......................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..TCCAGCCAGGTGACCGTC....................................................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ........................................................GGCAGGGGCCAACCAGGTCGC.............................................................................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .............................................................................................................................................................................CAAGGGAAGGGGAGTGC............................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| .................................................................................................................................................................CCTCCTCCTGGTCAGGGGCA...................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
|
GTGGGTCGGTCCAGTGGCAGAGGTCGTCGGTCGGTCCAGCGGAGGAGGTCCAATCACCGTCCCCGGTTGGTCCAGCGGTGGAGGTCCAGTCCCCGTCATCGGTCGGTCCAGTGGGAGAGGTCGTGGTCGGTCCAGTGGAGGAGGTCGTCGGTCGGTCCAGTGGAGGAGGACCAGTCCCCGTCCCCTCACGTGGAGGAGGTCAGCGGCCGGTAAGGCCGGGCAGAGGGGCGGCTCCCCTTTCACGGACTCCC
**********************************************************************(((..((.....))..)))(((.((((((((..(((((..((.(((.....))).)))))))...((....)).((((.((((......((.....)).)))).))))...)))))..)))))).******************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M058 embryo |
M044 female body |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................CGGTCGGTCCAGTGGAGGA.................................................................................... | 19 | 0 | 20 | 4.80 | 96 | 96 | 0 | 0 | 0 |
| ....................................................................................................................................................CGGTCGGTCCAGTGGAGGAG................................................................................... | 20 | 0 | 20 | 1.05 | 21 | 21 | 0 | 0 | 0 |
| ....................................................................................................................................CAGTGGAGGAGGTCGTCGGTC.................................................................................................. | 21 | 0 | 20 | 0.70 | 14 | 0 | 0 | 14 | 0 |
| ...............................................................CGGTTGGTCCAGCGGTGGA......................................................................................................................................................................... | 19 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| .............................................................CCCGGTTGGTCCAGCGGTGTA......................................................................................................................................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| .......GGTCCAGTGGCAGAGGTCGTC............................................................................................................................................................................................................................... | 21 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................CGGCTCCCCTTTCACGGACTC.. | 21 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 |
| ..........................................................................................................................................AGGAGGTCGTCGGTCGGTCCAGTG......................................................................................... | 24 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 |
| .....TCGGTCCAGTGGCAGAGGTC.................................................................................................................................................................................................................................. | 20 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................GAGGAGGTCGTCGGTCGGTCCAGTGG........................................................................................ | 26 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GCGGCTCCCCTTTCACGGACT... | 21 | 0 | 20 | 0.15 | 3 | 0 | 0 | 3 | 0 |
| ........................................................CCGTCCCCGGTTGGTCCAGCG.............................................................................................................................................................................. | 21 | 0 | 20 | 0.15 | 3 | 2 | 0 | 1 | 0 |
| ........................................................CCGTCCCCGGTTGGTCCAGC............................................................................................................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 |
| ........TTCCAGTGGCAGACGTCG................................................................................................................................................................................................................................. | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................GAGGAGGTCAGCGGCCGGT........................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................GTGGAGGAGGTCAGCGGCCG.......................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................GGAGGTCAGCGACCGGTAAGG.................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................CGGTCGGTCCAGTGGGCGA..................................................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................CCAGTGGGAGAGGTCGTCG............................................................................................................................. | 19 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TGCGGCTCCCCTTTCACGGAC.... | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................GGCTCCCCTTTCCCGGACTCC. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................................GAGGGGCGGCTCCCCTTTCACGGACTC.. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......................................CGGAGGAGGTCCAATCACC................................................................................................................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .......................TCGTCGGTCGGTCCAGCGGAGGA............................................................................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................TGGTCGGTCCAGTGGAGGA............................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................TTTGGAGGTCCAGTCCCCGTC.......................................................................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........................................................CAGTCCCCGGTTGGTCCACCG.............................................................................................................................................................................. | 21 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................GGTCGTCGGTCGGTCCAGTGGA....................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................CGGTCCAGTGGAGGAGGAC................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ........................................................CCGTCCCCGGTTGGTCCATCG.............................................................................................................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GGCTCCCCTTTCACGGACTCC. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................GTTCCCGGTTGGTCCTGCG.............................................................................................................................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................CGTCGGTCGGTCAAGTGGAGG..................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................................GGTCCAGTGGGAGAGGTCGTC.............................................................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:40750-41000 - | dan_1440 | CACCCAGCCAGGTCACCGTCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGTGGCAGGGGCCAACCAGGTCGCCACCTCCAGGTCAGGGGCAGTAGCCAGCCAGGTCACCCTCTCCAGCACCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGGTCACCTCCTCCTGGTCAGGGGCAGGGGAGTGCACCTCCTCCAGTCGCCGGCCATTCCGGCCCGTCTCCCCGCCGAGGGGAAAGTGCCTGAGGG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:52 AM