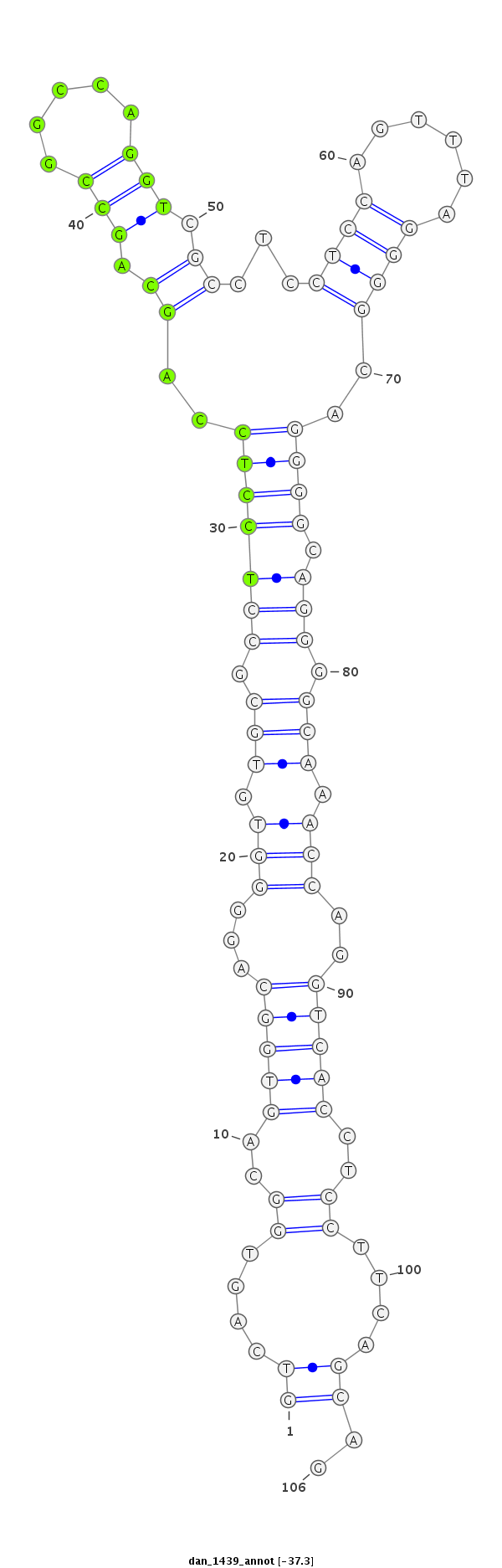
ID:dan_1439 |
Coordinate:scaffold_13150:41156-41261 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
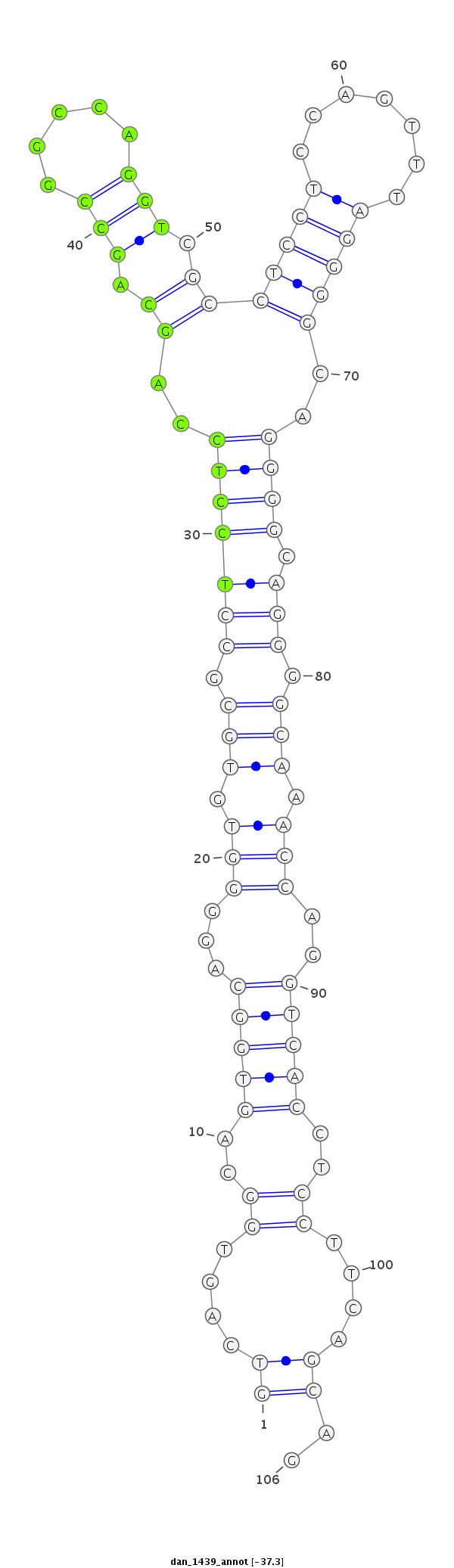
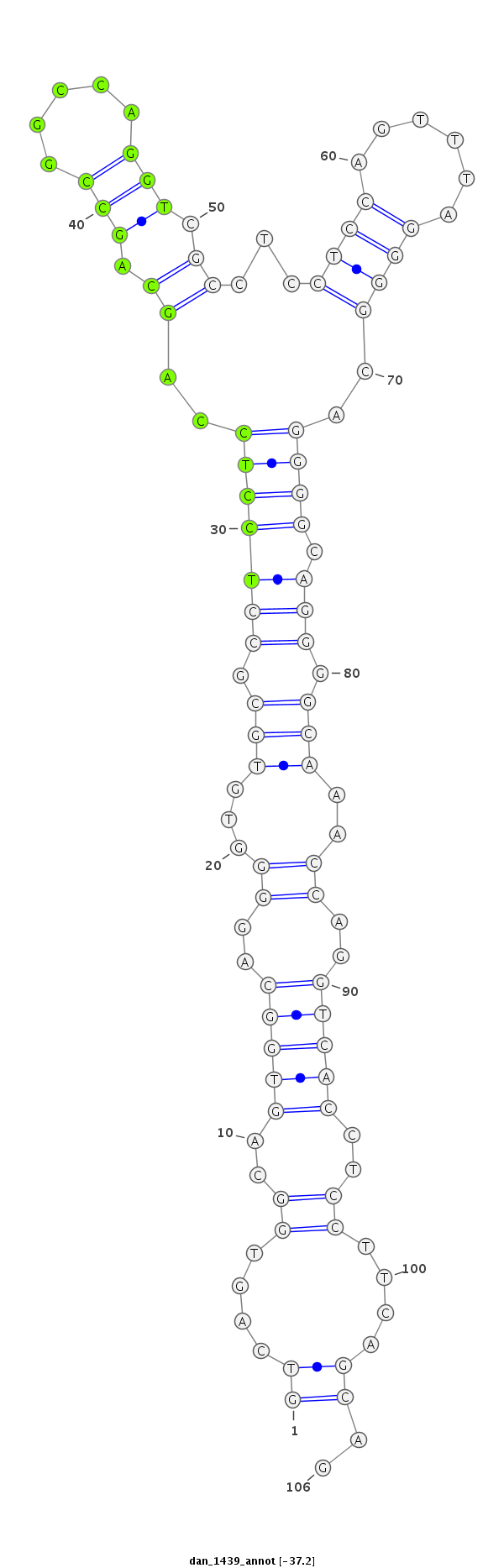
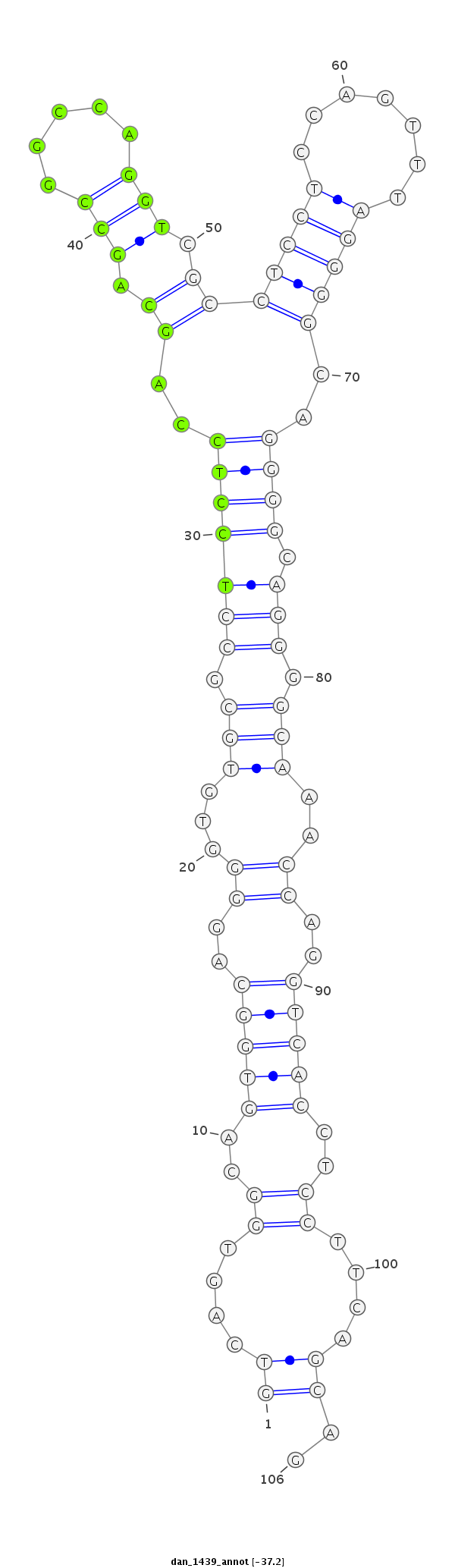
| -37.3 | -37.2 | -37.2 |
 |
 |
 |
intron [Dana\GF18978-in]; exon [dana_GLEANR_20368:2]; CDS [Dana\GF18978-cds]; CDS [Dana\GF18978-cds]; exon [dana_GLEANR_20368:3]; intron [Dana\GF18978-in]
No Repeatable elements found
| ----------########################################----------------------------------------------------------------------------------------------------------################################################## CCTCCTGCAGGGACAGGGCAAGTTGGGTGCGGCCATATCGCCTCCTCCAGGTCAGTGGCAGTGGCAGGGGTGTGCGCCTCCTCCAGCAGCCGGCCAGGTCGCCTCCTCCAGTTTAGGGGCAGGGGCAGGGGCAAACCAGGTCACCTCCTTCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGGAGCAGGGGCCAGCCAGGTCGCC **************************************************((....((..(((((...(((.(((.(((((((..((.(((.....))).))...((((......))))..)))).))).))).)))..)))))..))....))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V039 embryo |
M044 female body |
M058 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| ...................................................TCATTGGCAATGGCAAGGGTGTG.................................................................................................................................... | 23 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................TCCTCCAGCAGCCGGCCAGGT........................................................................................................... | 21 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 |
| .........................................................GCAGTGGCAGGGGGGGGCGC................................................................................................................................. | 20 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 |
| .......CATGGACAGGGCAAGTAG..................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 |
| .................................................................................................GTCGCTTCCTCCAGTTTAGGGG....................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................TTCACCAGCCAGCCAGGTCGC..................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................GAGCACGGGCCAGCCAGGCCG.. | 21 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................GTGCGCCTCCTCCAGCACCCG.................................................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ....................................ATCGCCTCCCCCGGGTGAGTG..................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
GGAGGACGTCCCTGTCCCGTTCAACCCACGCCGGTATAGCGGAGGAGGTCCAGTCACCGTCACCGTCCCCACACGCGGAGGAGGTCGTCGGCCGGTCCAGCGGAGGAGGTCAAATCCCCGTCCCCGTCCCCGTTTGGTCCAGTGGAGGAAGTCGTCGGTCGGTCCAGCGGAGGAGGTCCAATCCTCGTCCCCGGTCGGTCCAGCGG
**************************************************((....((..(((((...(((.(((.(((((((..((.(((.....))).))...((((......))))..)))).))).))).)))..)))))..))....))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
M044 female body |
M058 embryo |
V106 head |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ........CCCCTGTCCCGCTCGACCCACG................................................................................................................................................................................ | 22 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...TTACGTCCCTGTCCCGTTCTA...................................................................................................................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...TGACGACCCTGTCCCGTTCTA...................................................................................................................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................TCCAGCGGAGGAGGTCAAAGC.......................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................GGAGGAGGTCAAATCCCCGTC.................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................CGGTCCAGCGGAGGAGGTCAA............................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................CCCGTCCCCGTTAGGTCCAGT............................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................TGGAGGAAGTCGTCGGTCG............................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................CCACACGCGGAGGAGGTCGTC..................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................CGGAGGAGGTCCAGTCACTGT.................................................................................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................GGTCCAGTGGAGGAAGTCGTCG................................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TTTCGTCCCCGGTCGGTCCAGC.. | 22 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................TCGGTCGGTCCAGCGGAGGAGGTC............................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................CACGCGGAGGAGGTCGTCG.................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................CGGAGGAGGTCCAATCCTCGT.................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................TTGCGGAGGAGGTCCAGTC....................................................................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:41106-41311 - | dan_1439 | CCTCCTGCAGGGACAGGGCAAGTTGGGTGCGGCCATATCGCCTCCTCCAGGTCAGTGGCAGTGGCAGGGGTGTGCGCCTCCTCCAGCAGCCGGCCAGGTCGCCTCCTCCAGTTTAGGGGCAGGGGCAGGGGCAAACCAGGTCACCTCCTTCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGGAGCAGGGGCCAGCCAGGTCGCC |
| Species | Read alignment | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:49 AM