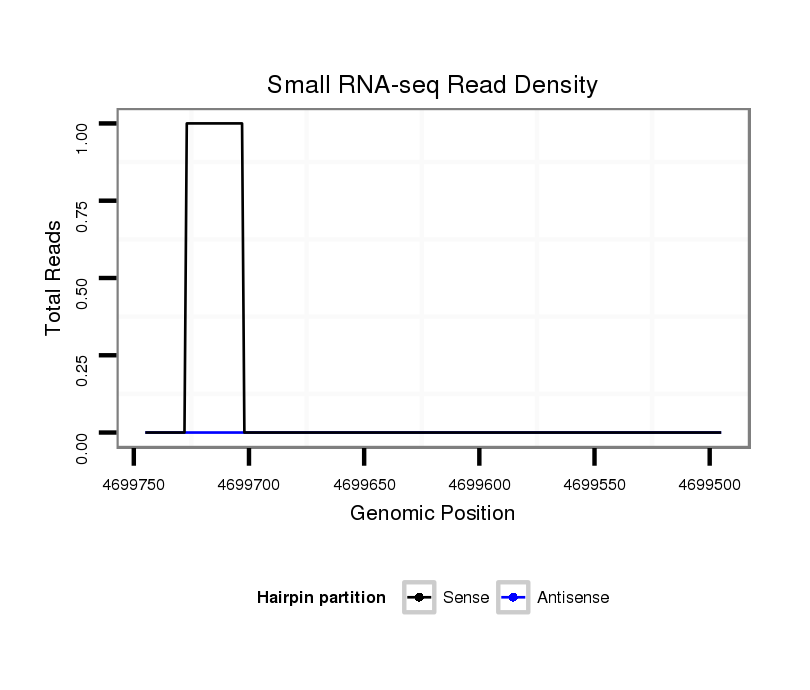
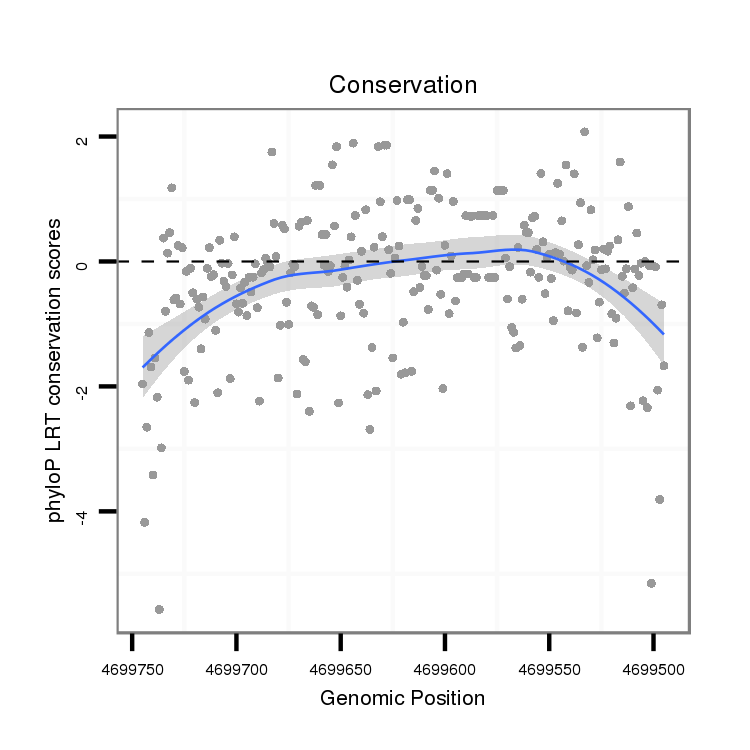
| droAna3 |
scaffold_13117:4699495-4699745 - |
dan_1387 |
GACAGCCGGC-GGCAAGCA---CTTGTTCCATGCACCAAT--------------------------------------------------------------------ACCCGT-----GC---------------------AGGCAGGTTTTCCTTTGATTTA-CTTTTTTTGCTTTTA----------------------------------------------------------------------------------------------------------------------------------------------------------CCACAAAAACATTA--------TCCCAAAAATA-A--AAATTA-------------------------------------------------------------------------------------AAAAAAGAA---AA-------------------------CTC------------------------------------------------------------------------------------------------------------AAAAAAAGGGAAA--------------------------------TTGTTAA----------------------------------------------------------------------------------GTTTAAAATAGCTTATATAATTATATTTCAGTCTT-----AAATATTTTAAA------------------------------------------------------------------------------------------------------------------AACTATTT----------------CAAAAACTTG-TTCAGATCTTACAA-----------------------------------------------------CTCTTCA--AATAA-T--TTTTTCA-AATTA |
| droBip1 |
scf7180000395814:6587-6814 - |
|
CAGG-----C-GGCAAGCACTCCTTGTTCTATGCACCAAT--------------------------------------------------------------------ACCAGT-----GC---------------------AGGCAGGTTTTCCTTTGATTTTACTTTTTTTGCTTTTA--A------------------------------------------------------------------------------------------------------------------------------------------------------------AAAACATTT--------TCCCAAAAAAA-G--AA---A-------------------------------------------------------------------------------------AAAAAAGAA---AA-TCCCAAAACGGAGGAAAAGGAAAT---------------------------------------------------------------------------------A-------------TTAAGAATATATACCT--------------------------------------------------------------------TTAAA------AAGTTATAGATATAAAGCTATA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGTCCAATAC----TA-------------------------------------TT----TTTT-------------------------AAAATTT------------------------TTAA-TATTTTTTAA-ATATT |
| droKik1 |
scf7180000302696:915155-915421 - |
|
AGCCGCCGTT-GGCAAGCA---CTTGGTCCATGCACCAAT--------------------------------------------------------------------CCCAGT-----GC---------------------AGGCAGGTTTTCCTTTGATTTTCTTTGCTTTTCTTTTC--C------------------------------------ATTACAATTGGCCCC-------------------------------------CAGGGCAG-------------------------------------------------------------------TG--------GCCGGG-------------------------------------------------------------------------------------------------------------------------------------------------AAATGCCAGGGAAACGAACTCAAAAGTCCAAGAACTTGATTGTAACGTAAGGGAGATATTCATTAAAC---------AT------------------------TT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------TG-----GAATATTTTAAAAATATAGACCATTATTATT-----------------------------------------GAAA------------------------GGTT-------------------------------------------------------C-TTAA--GTTTACAT-----------------------------------------------------TTCTTAA--AGAAAGA--TAAATAATATTTT |
| droFic1 |
scf7180000453906:183012-183273 - |
|
GACACGCAGTGGACAAGG------GACCACAATAG------------------------------------ACTTTTGAGGG---------------------------CC----------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAAGTCCAGAGATGCCAGCGTGAACGTGAGT------------TCATTGGCCGTGAATAAAAAACCATTC--------TCCTAAAATGA-G--AATATA-------------------------------------------------------------------------------------AAAAAAGTA---AA-------------------------CAT----------------------------------------------------------------------------------------------------ATATAGCA-----ATAAATAATTAAAAGGAGTGA---------------------------ATAAAATAGACATAA---------------------------------------------------A----------------------------------------------------------------------------------------TATAATGAATAATGCAATTACTATT-------------AAAA-----------------------CAGCTCTTAAGTACTTACA--------------------------------------------------------TACTTGTT-------------------------GCATCTGTA-------------A-------ACAGA-C--TTTTTCA-AATAT |
| droEle1 |
scf7180000491194:467846-468057 - |
|
AGAAGATCCA-AGTT---------TGTAAT---------------------------------------------------------------------------------------ACTC---------------------AAAGTAATTTTCGTTTGATTGGAGTTTTTTTAATTTTT--T------------------------------------------------------------------A----------------------------------------------------------------------------------------------------------ATTTAA-------AAAT-CTG--------------------------------------------------------------------------------------AAAACCTA---AA-------------------------TTT-----------------------------------------------------------------------TAACCAAATTAAA-----------------------------------------AAGGACTCA---------------------------TTTAAAATCATATAT---------------------------------------------------TTTTTACATTAGT-----------------------------------TC-----AAATATTTT--------------------------------------------------------------------------------------------------------------------------------ATATTAATTCTTTTAGAAATA---TACA---------G------------------------TTTTGAATTCTACAG--------------CC-------ATTAA-T--TTATTTT-AACTG |
| droRho1 |
scf7180000769211:15121-15330 - |
|
ACTACAAGTA-GAAAAGC------AGCAATG--CGTCA----------GCTC------------AT-----------GAAAACAACCCACTCGAT-GCCTGCGCCCACCCCAAA-----A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACCATATTC--------TTTTACAGT-G-G--ACGCTC-------------------------------------------------------------------------------------AGAAAGGAG---AA-------------------------CTT----------------------------------------------------------------------------------------------AGAAAAGTAACATT--------------------------------------------------------------------TTAAA------TAGTTTAAGCTTTAGAAATATCAAACAGTTTTTCATTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAA----TAGATTTTATTTTTATATA--AAAAG---GCATTTCA-AATTT |
| droBia1 |
scf7180000302188:2034806-2035061 + |
|
GATGGCCAGC-GA--ACAA---CCTGTTACATCGCCCGCT--------------------------------------------------------------------GCCTGAAATATGTAAAGTCAATTTCGTGTATCGCA-TCG-GAAAACTGTA---------------ACTAATG--C------------------------------------------------------------------------------------------------------------------------------------------------------------CAAACATTAACCCATTAACGCGA-------AAAC-CAG--------------------------------------------------------------------------------------AGAAATCG---G--------------------------C------------------------CAAAGAGC---------------------------------------------CAGACGAAAATGGGGTACCAGC------------------------------------------------------------CAA--------GCATAAATTTCAC---------------------------------------------------TAATTAGCAGTA---------------------TATTACA-ATTT---------------------------------------------------------------------------------AAGTATGCAAGATCTTTT--------------------------------------------------------TAGGAACTTATTTCAGATCTTTAAA-----------------------------------------------------CT----------------TCTAAGT-ACTTT |
| droTak1 |
scf7180000414375:281158-281355 - |
|
CGATGGC-------AAGCA---ATCGGTGTACACACCTCC--------------------------------------------------------------------TCCAGT-----GC---------------------AGGCAGGTTTTCCTTTGATTTTCTTTGCTTTCCTCCTCCCC------------------------------------ATCAAAAAAGGGCCA-------------------------------------ACGGAAGA---------------------------------------------------------------------------------------------T----------------------------------------------------------------------------------------------------GG----------------------------CAGGAAACGCTGGGGAAACGAA----------------------------------------------------------------------------------AATTG-------------------------------------------------------------------------------------------------------------------------------------------------------------TATTTCCTTTTG-----GGATATTCTAGA------------------------------------------------------------------------------------------------------------------AATCGTTT----------------T-------------------------------------------------------------------------ATTCTAATATTAAAATAA-TAATTTCTCA-ACTTA |
| droEug1 |
scf7180000409908:13424-13612 - |
|
AACAACAAGG-AGC-AGCA--GCTAAC----------------------------------------------------------------------------------TCAAT-----TC---------------------AAAAAAAATTTTTTAT-------------------------ACATATACATATACTTATTTAATTACGC--------------ACATACCT----------------------------------------------------------------------------------------------------------------ATA--------TTATAAAAAAA-A--ACATTA-------------------------------------------------------------------------------------AAA-----A---AA---CGCAAAGGTTGGCAATAACAAT---------------------------------------------------------------------------------A--------------------------------------------------------------------------TT-----------------------------------------------------------------------------------------------------------------------------------------------------------GTT-----------------------------------------AACA-----------------------GATTTTTTAAGTTGCTATATAGATAAAATA----TT-------------------------------------------TTCC-------------------------GAAA-----------------------------A----------AA-CGATT |
| dm3 |
chrX:8713981-8714188 + |
|
GAACAGAGC--GACCAACA-----TAATA-------------------------------------------------------TCATCTACAAT-AGTTATGAGCATCCAATT-----TC---------------------AGGCAAATATTA-TATAATATTACTTGCGTTGCTCATC------------------------------------------ACAATATAC---------------------------AGCATAAGAAACCGCACAAGG---------------------------------------------------------------------------------------------T----------------------------------------------------------------------------------------------------AA-CT---------------------------TAA----------------------------------------------------------------------------GAAACTTAAGAAAAAGT-----AAATAG--------------------------------------------------------------------TTTAA------TTGCCTATATT-----------------------------------------------------------------------TGAGCAATATAAAAAATCTTTT---------------------------------------------------------------------------------------------------------------------------------------------AAAA--------------------------------------------------------------------------------------AA---TCTACAAA-AAACT |
| droSim2 |
3r:9408384-9408599 - |
|
GCGAGCCGGA-GG--AGCA---GT----ACG----------------------------TGGTGGTGCAACGCATCCAGCGC-C-----GAATCCTC-----------ACC-----------------------------ACA-GCGGGCACACTGTAAGTAACCTCTGTAGACCTTCTTTCCCTGAAAACATAAAC------------------------------------A-------------------------------------TC-------------------------------------------------------------------------------------ATAAAATTG-C--ATTTTA-------------------------------------------------------------------------------------AATAAAATC---AT-------------------------CTA------------------------------------------------------------------------------------------------------------AAGCAAAAGGCAA--------------------------------TCGAT----------------------------------------------------------------------------------------------------------------------------------AGA--TAATATTTACCATTATTATTAAATAG----TATTA-------------TATTATTAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-----AA---CA-TTTCA |
| droSec2 |
scaffold_711:9280-9488 + |
|
AAAAGCCGTC-GGCAAAT------CGCAAAACTCG---AA------------AATTAAAA--------------------------------------------------------------------------------------------TTAGGA-------------------------ACATGTGAGTAGGCTTTTTGTTCTAAGTTTTCGCCAATCAAAACATATCCAGTGAATAAATTTTTTC---------------------------------------------------------------------------------------------------------------------------GTTT-------------------------------------------------------------------------------------AAA-----A---AA-------------------------AGTAAAAAATACTG-------------------------------------------------------------------------------------------------AAA--------AAAAAT----------------------ATAACATC--CAAGCAATTTTGGAA--------------------------------------------------------------------------------------------------------------------------------------------------------TTGC-------------TTGGAATATT---------------------------------------------AAAA--------------------------------------------------------TATTTTTC-------------------------TCGTCT--------------------------------GTGTTT----TAAA |
| droYak3 |
v2_chrUn_6924:7441-7635 + |
|
ACAAGAATT---------------TTCTGTTTATACAAACACACATATATAT------------AT-----------AAAT---ACATATACAAT-ACATACACACGTACTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATATATTAAGGACTTACAATCAAATTAAGCGCATCTATAAAAATAAAAAAAAAAA---AA-------------------------TTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATGATGTCAGAAA-----------------------------------------------------------------------------------------------------------------------------A----GACTA-------------CTTAATTCCTTAA------------------------------------------AATGCAGTCCGAAAT----TA---------------------------------------------------------------------------------------------------------------TTTTGGA-AACTA |
| droEre2 |
scaffold_4929:22027084-22027280 + |
|
ACAAGAATT---------------TTCTGTTTATACAAACACACATATATAC------------AT-----------AAAT---ACATATACAAT-ACATACACACGTACTATT-----AA---------------------GA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTACAATAAAATCAAACCCATCTATAAAAAAAAAAAAAAAAAAAAA-A----------------------------AAA----------------------------------------------------------------------------------------------------AATT--------------------------------------------------------------------TTAAATGATGTCAGAAA-----------------------------------------------------------------------------------------------------------------------------A----GACTA-------------TTCAATTCCTTAA------------------------------------------GAAGCAGTCCGAAAT----TA---------------------------------------------------------------------------------------------------------------TTTTGGA-AATTA |
| dp5 |
XR_group6:9122499-9122731 + |
|
AACAACAACA--GC---------------------------------------------------------------------------------------------------------AA---------------------CAACAACTATTCTTATGGTTTTCTTGATTAGGCAATTT------------------------------------------------------------------TATAATTTAAACAGGAAGAAAAGAAGCAGAAGA---------------------------------------------------------------------------------------------A----------------------------------------------------------------------------------------------------AA-GA---------------------------AAC----------------------------------------------------------------------------AAA---------------------------------------------------------------------------------------------ACAAAAGAAAACAGAAGATGCAGAAAAAAGAACTAATAAGAAATCATTAAATATACACGTAATTAT-------------------------------------------TAATTAAAC------------------------------------------------------------------------------------------------------------------AATAATTA----------------TATAGAATTA-TACACATAATACA----CCAT-------------------------ACAACTGTT---------------------AGTAG-T--TTCCTTT-TTTTT |
| droPer2 |
scaffold_0:8268152-8268267 - |
|
ATCAAAAAAA--C---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCAAAAAAAAA--GTTTTA-------------------------------------------------------------------------------------AAAAAAGAA---AA-------------------------TCTAAAAAAA-------------------------------------------------------------------------------------------------AATTAAT--------AAAATT----------------------ATAGAATC--GTAAAAAAATTAGAC--------------------------------------------------------------------------------------------------------------------------------------------------------ACAA-------------TTAAACTACTTAA------------------------------------------AAAAA-------------------------------------------------------------------------------------------------------------------------------GATAT----AAAA |
| droWil2 |
scf2_1100000004521:4407132-4407381 - |
|
AATAACCAAGCGACAACA------AG-----CATGTT----------------------------------ACTTTTGCAGC---------------------------CCAAT-----GT---------------------AAGTTGATCGAAATTTAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATGAAAATTTCTTAGAA---------AA------------------------TT--------------------------------------------------------------------TCTAA------TTAGTTAAAAC-----------------------------------------------------------------------AAAACAAAAAAAAAAACCTTTC--TAAAATTAGCCAACATTGTTAAGTAGAGCCAACTATGCAA------------------------A------------------------TGTT-------------------------------------------------------A-TTAA--GTCATCAT---TATTGCATAAGAAATACCAAACTTTAAT-TAAAC----TTGATCATAAAATTCTCTC--GATTA---GTGTGTTT-AAACT |
| droVir3 |
scaffold_12963:8991218-8991417 + |
dvi_11046 |
AGTCATTC-A-GGTAAATG---TC----ACGTA--CCGAA--------------------------------------------------------------------ACAC-------TC------AAT----------T----------------TA--------------GCTGGAA----------------------------------------------------------------------------------------------------------------------------------------------------------CAACAATAATATTT--------AACCAAAAAAA-A--AA---A-------------------------------------------------------------------------------------AAA-----A---AA-------------------------AAC--------------------------------------------------------------------------------AAA------------------------A-----------------CAGAACAAATTAAAAACCAAAAC------------------------------------------------------------------------------------TCATTGCCATTATT-----------------------TCTGCGTT---------------------------------------------------------------------------------AATTGTTTACCATTTCTTTCATTAATTT-------------------------------------------------------T-TTAA--ACTAACAA-----------------------------------------------------ACCTCCA--AGCTATA--TTTTTC---TAAT |
| droMoj3 |
scaffold_6496:6986974-6987251 + |
|
AACGTCTGGC---CAGGCA---ATTGTTAGGCTCGACAGA--------------------------------------------------------------------AGCTGC-----AC---------------------AGTGGGGCAA-GTGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGAGAGATTAGGCTTACATACAAAGCTTACATACAAGCAAAGATTATTGAAGAGT-----------------------------------------------------------------------------AAAAGCTGTTGAAAGAA-CTTCAAAATAAAATAAATAAATAGAAAATA-ATAAA--TAA---------TAATAAAA-----------------------------------------------------------------------------------------------------------TAAATAA-----------------------------------------------------------------------------------------------------------------------------A----AAATA-------------ATATAAAAATAAT------------------------------------------AAAACAAATAAAAAA----TATTATAAAAACAAATATAACTATT-G-TCAA--GTCTTCTT---TCC--------------------------TAATTT------------------------AATAA-T--TTTAT----TAAA |
| droGri2 |
scaffold_15074:2135032-2135326 + |
|
GGTACCAG-C-GGCAAGCG---CTGG----ACA-----GG---------AG-CAAAAAGGTGAGGCTTAATGACTTTGCCAT-C-----AAGCATAC-----------G--------------------------------------------CCATGGGTTTA-CTTTGA------------------------------------------------------------------------------G-----------------------------------------------------GCAAATCAATTAGTTAAAACTATGACTGCATTCAAAATACCATTA--------TCCTTAAATAA-A--AT-CTG--------------------------------------------------------------------------------------AAAAAGCA---AG-------------------------CCT-----------------------------------------------------------------------TC---------A------------------------C-----------------TTGAAAAAATTATTCACTAAAAGTCAAATCGCAAA----------------------------------------------------------------------------------GATGTTTGTGGCAAATATATATTTATTTTTTTCTT-----TACCTTTTCAAA------------------------------------------------------------------------------------------------------------------TTTGATGTCTTATATTAGAAG--------------TTTCGGTCTTACAT-----------------------------------------------------CT-----------------TTGAAA-TATTT |