ID:dan_1327 |
Coordinate:scaffold_13117:2983519-2983564 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
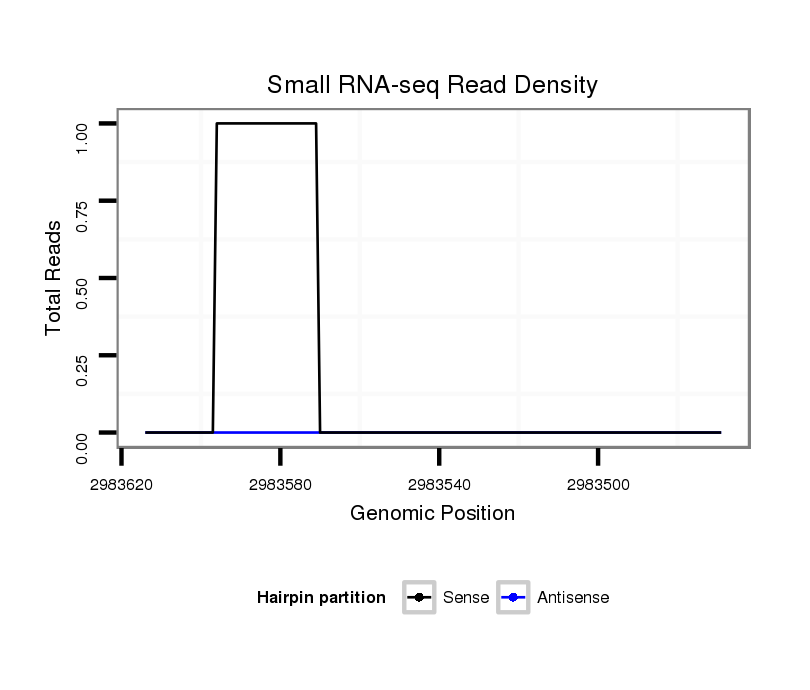
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
intron [Dana\GF20929-in]; exon [dana_GLEANR_4169:2]; exon [dana_GLEANR_4169:1]; CDS [Dana\GF20929-cds]; CDS [Dana\GF20929-cds]
No Repeatable elements found
| ##################################################----------------------------------------------################################################## CTGCCACCATCACAAACATGATACCATCTACCGCAGACGTCCCAGGTCAAGTAAGTGTCTCTTAAACCATTCTAAGGAATGGTTTTAACCTTTCAGTTTTTAGTGGAGGTGCCGCAAGAAGCTCAGCCAGTCTTGCCAACCACTAC **************************************************..(((.((.....(((((((((....)))))))))..)))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
M058 embryo |
V105 male body |
V039 embryo |
M044 female body |
|---|---|---|---|---|---|---|---|---|---|---|
| ....CACCAACACATATATGATACCATC...................................................................................................................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................TGATACCATCTACCGCAGACGTCCCA...................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................TAGGGCAAACGTCCCAGGTC.................................................................................................. | 20 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................TATAACGCAGACGTCCGAGG.................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........TAACAGACATGATACCTTCT..................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................GCAAGTATGGGTCTCTTAAA................................................................................ | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................TGTTTCTTAAACCACTCTAC....................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................TCAGTATTGCTAACCACTA. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
GACGGTGGTAGTGTTTGTACTATGGTAGATGGCGTCTGCAGGGTCCAGTTCATTCACAGAGAATTTGGTAAGATTCCTTACCAAAATTGGAAAGTCAAAAATCACCTCCACGGCGTTCTTCGAGTCGGTCAGAACGGTTGGTGATG
**************************************************..(((.((.....(((((((((....)))))))))..)))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V106 head |
V055 head |
M044 female body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................ACCACAGAGAATTTGGTAA........................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................TACTATGGTAAATGGCGTACG............................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................TACAATGGTAAATGGCGT............................................................................................................... | 18 | 2 | 8 | 0.25 | 2 | 0 | 0 | 0 | 2 | 0 |
| .......................................................ACAAAGAATTTGGTGAGA......................................................................... | 18 | 2 | 10 | 0.20 | 2 | 0 | 2 | 0 | 0 | 0 |
| ......................................................AACAGAGAATTTGGTGAGA......................................................................... | 19 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................CGGTCAGAACGGGAGGTTA.. | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 1 | 0 | 0 |
| .....................................................TCACAAAGAATTTGGAGAGA......................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................AGAATTGGACAGTCATAAA............................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................CAAAGAATTTGGTGAGA......................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................CAAAAGTGGAAAGACAAAAA............................................. | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................AGGTCAGAACGGGTGGT.... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13117:2983469-2983614 - | dan_1327 | CTGCCACCATCACAAACATGATACCATCTACCGCAGACGTCCCAGGTCAAGTAAGTGTCTCTTAAACCATTCTAAGGAATGGTTTTAACCTTTCAGTTTTTAGTGGAGGTGCCGCAAGAAGCTCAGCCAGTCTTGCCAAC--CACTAC |
| droFic1 | scf7180000453882:1131902-1131905 + | CC------------------------------------------------------------------------------------------------------------------------------------------------AC | |
| droEle1 | scf7180000491268:461028-461043 - | GTCT---------------------------------------------------------------------------------------------------------------------------------TGGCATC--C-ACAT | |
| droRho1 | scf7180000771936:3751-3803 + | CCGT----------------------------------------------------------------------------------------TCTGTTCCTGGAGGAAACGTGGCAAGACG-----CCATCGATGCCATG--ACCAAT | |
| droYak3 | 3L:17675928-17675929 - | A--------------------------------------------------------------------------------------------------------------------------------------------------C | |
| droEre2 | scaffold_4845:6148399-6148408 + | CAACTA------------------------------------------------------------------------------------------------------------------------------------------CCGC | |
| dp5 | XL_group1a:7547436-7547462 + | CTCA-------------------------------------------------------------------------------------------------------------------------ACCAATCTTTACAAATTGAACAC | |
| droPer2 | scaffold_2:2858842-2858847 + | CGC----------------------------------------------------------------------------------------------------------------------------------------------CAC |
| Species | Read alignment | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 01:17 AM