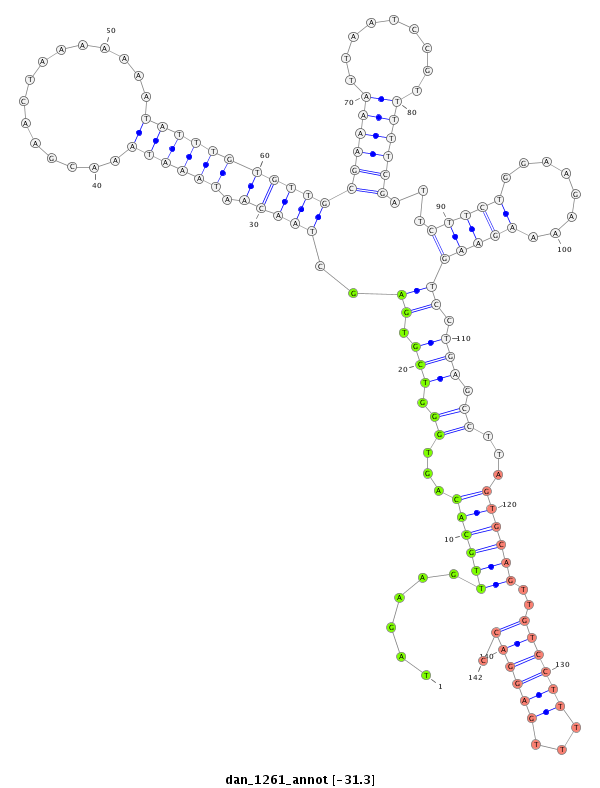
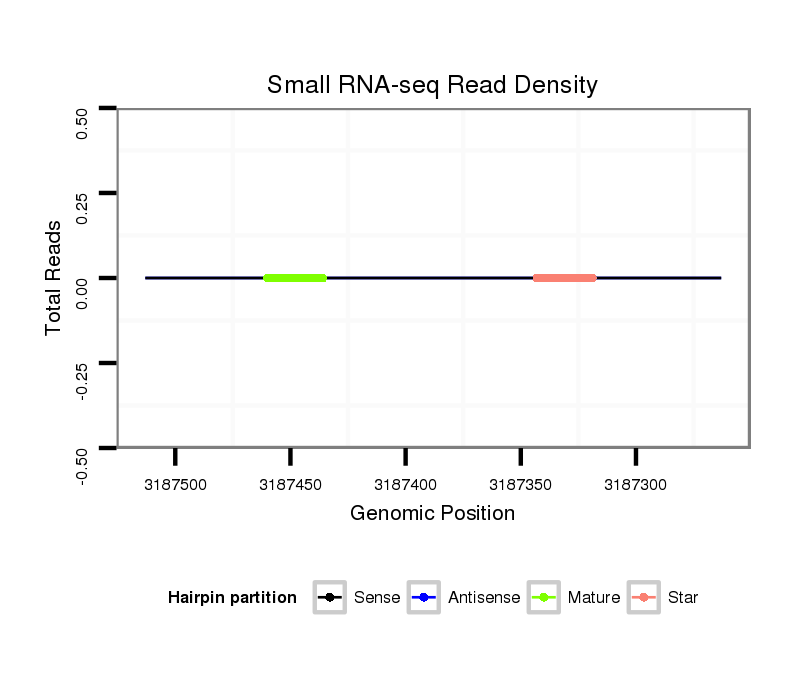
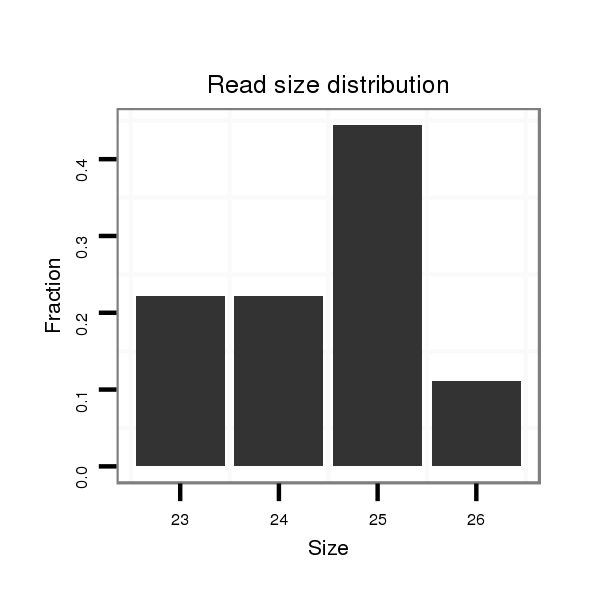
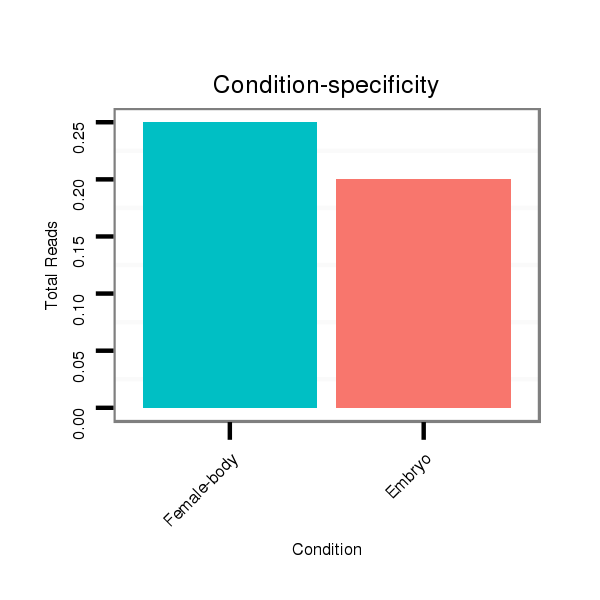
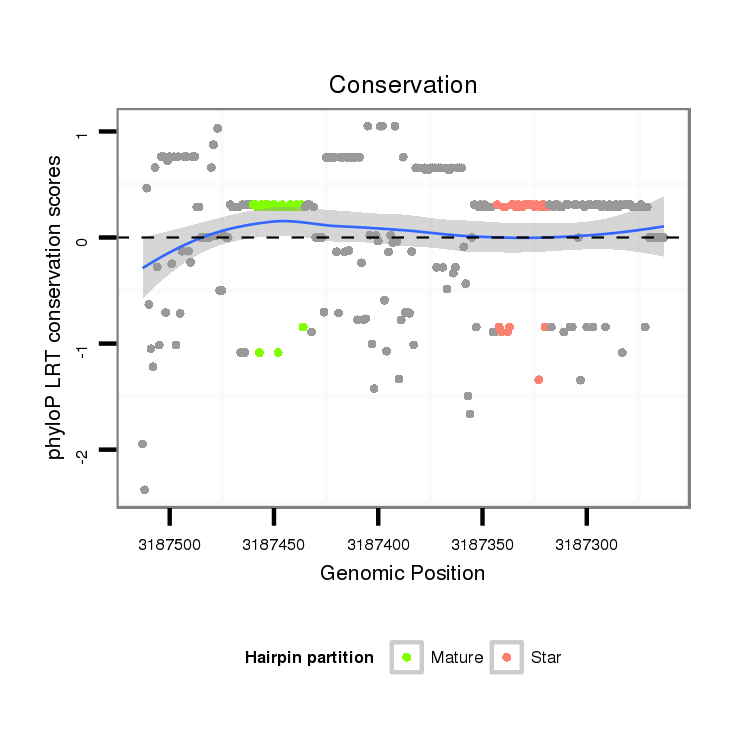
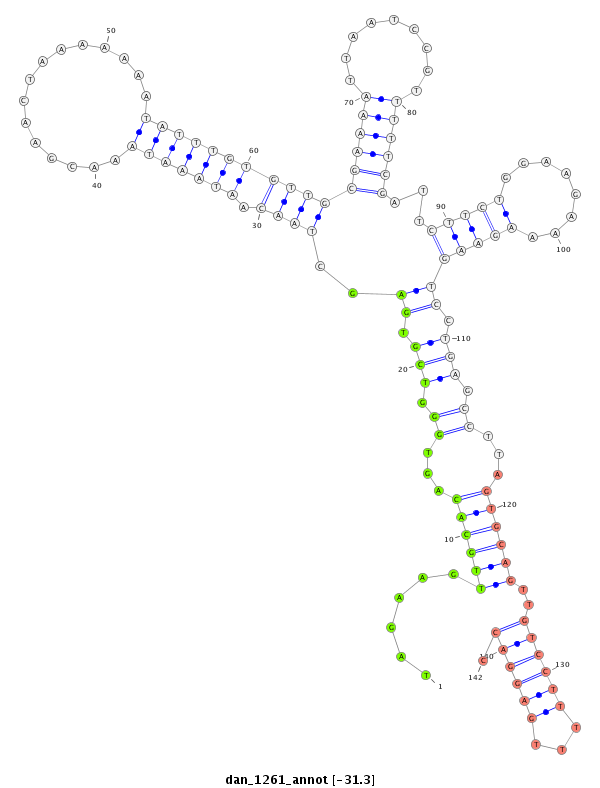
ID:dan_1261 |
Coordinate:scaffold_13099:3187313-3187463 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.3 | -30.6 | -30.6 |
 |
 |
 |
CDS [Dana\GF22791-cds]; exon [dana_GLEANR_7134:2]; intron [Dana\GF22791-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATACGCTGGTTACAGAGTGCTACAATCGCTCATACATATATTACCCCGTGATATAGAAGTTGCACAGTGGGTCGTGAGCTAACAATAAATAAACGAACTAAAAAAATATTTGTGTTGCGAAAATTAATCCGTTTTTCGATTCTTCTGGAAGAAAAGAAGTCCTGAGCCTTAGTGCAGTTGTCCTTTTTGAGGACCTTCAAGGTTTTTTTCCTCCTTGAAAGAAGTGGAGAAATTTCCGGAGGCGATGATAA *****************************************************......((((((...((.(((.((..(((((.((((((...............)))))))))))((((((.........))))))...(((((........))))))).))).))...))))))..((((((...)))))).******************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
|---|---|---|---|---|---|---|---|
| .....................................................TAGAAGTTGCACAGTGGGTCGTGAG............................................................................................................................................................................. | 25 | 0 | 20 | 0.15 | 3 | 0 | 3 |
| ....................TACAATCGCTCATACATATATTACC.............................................................................................................................................................................................................. | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 |
| ....................TACAATCGCTCATACATATATTACCCCG........................................................................................................................................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 2 | 0 |
| .....................................................TAGAAGTTGCACAGTGGGTCGTGAGC............................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ..........................................................................................................................................................................AGTGCAGTTGTCCTTTTTGAGGACC........................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ....................TACAATCGCTCATACATATATTACCCC............................................................................................................................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| ....................................................ATAGAAGTTGCACAGTGGGTCGTG............................................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ...................................................TATGGAAGTTGCACAGTGGGTCGTG............................................................................................................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 1 |
| ....................................................ATAGAAGTTGCACAGTGGGTCGT................................................................................................................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| .....................................................TAGAAGTTGCACAGTGGGTCGTGA.............................................................................................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| .....................................................TAGAAGTTGCACAGTGGGTCGTG............................................................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| .................................................................................................................................................................................................................................GGAGAAATTTCCGGAGGCG....... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 |
|
TATGCGACCAATGTCTCACGATGTTAGCGAGTATGTATATAATGGGGCACTATATCTTCAACGTGTCACCCAGCACTCGATTGTTATTTATTTGCTTGATTTTTTTATAAACACAACGCTTTTAATTAGGCAAAAAGCTAAGAAGACCTTCTTTTCTTCAGGACTCGGAATCACGTCAACAGGAAAAACTCCTGGAAGTTCCAAAAAAAGGAGGAACTTTCTTCACCTCTTTAAAGGCCTCCGCTACTATT
********************************************************......((((((...((.(((.((..(((((.((((((...............)))))))))))((((((.........))))))...(((((........))))))).))).))...))))))..((((((...)))))).***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V039 embryo |
M058 embryo |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|
| .................................................CGATATCTTCAAGGTGTCA....................................................................................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ...................................................ATATCTTCAACGTGTCACCCAGCACT.............................................................................................................................................................................. | 26 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TTCACCTCTTTAAAGGCCTCCGCTACTA.. | 28 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TTCACCTCTTTAAAGGCCTCCGCTAC.... | 26 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................ATCACCTCTTTAAAGGCCTCCGCTACT... | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CAAGGAGGAATTTTATTCAC......................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...............TCACGATGTTAGCGAGTATGTATATAAT................................................................................................................................................................................................................ | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...................................................ATATCTTCAACGTGTCAGCCAGCACT.............................................................................................................................................................................. | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................ATGTTAGCGAGTATGTATATAATGG.............................................................................................................................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................CCTAGAAGTTCAAAAAAAA.......................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ..................................................TATATCTTCAACGTGTCAGCCAGCACT.............................................................................................................................................................................. | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13099:3187263-3187513 - | dan_1261 | ATACGC--------TGGTTACAGAGTGCTACAATCGCTCATACATATATTACCCCGTGATATAGAAGTTGCACAGTGGGTCGTGAGCTAACAATAAATAAACGAACTAAAAAAATATTTGTGTTGCG------------------------------------AAAATTAATCCGTTTTTCGATTCTTCTGGAAGAAAAGAAGTCCTGAGCCTTAGTGCAGTTGTCCTTTTTGAGGACCTTCAAGGTTTTTTTCCTCCTTGAAAGAAGTGGAGAAATTTCCGGAGGCGATGATAA |
| droBip1 | scf7180000393792:28889-29119 + | CGTTA-----------TTTGCAGATTACTACAATCG-------TT-----ACCCCTTTATATAGCAGTTGCACCGTGGGTCGTGAACTATC----GATAAACAAACTAAAATAAATTTATTGTTGAGAAAATGAAAATCCGTTTGTTGAGTCTGCATCTCCTGAAAGGAAGCTCT----------------------------TTCTGAGCCAAAAAGCTATTGTCCTTTTTGACGATCTCCAAGGATTCCTT-GTCTTTAAAAGAGGTGGAGACATTTCCGGAGAC-------- | |
| droRho1 | scf7180000772588:448-461 + | GGACGC------------------------------------CATGCT--------------------------------------------------------------------------------------------------------------------------------------------------------CC--------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000413255:274-287 + | GGACGC------------------------------------CATGCT--------------------------------------------------------------------------------------------------------------------------------------------------------CC--------------------------------------------------------------------------------------------- | |
| droYak3 | v2_chrUn_7001:1292-1334 + | TTACATATTATGTTTATTTACATATTATTATCAT-------ACATATT--------------------------------------------------------------------------------------------------------------------------------------------------------TT--------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_3334:92-147 - | AAAATA--------------------------------------------------------------------------------------------------------------TCCGATTTCTG------------------------------------ACAACAAATTTTTTTTTCGATTTTTTTTGATAAAAATAA--------------------------------------------------------------------------------------------- | |
| dp5 | Unknown_group_629:7018-7063 - | ACAAGA-----------------------------------------------------------------------------------------AATAAATAAATTGAAATACAATTTCTGTTACC------------------------------------GAAA-------------------------------GTAA--------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_12116:4052-4058 - | ATACAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
Generated: 05/18/2015 at 01:10 AM