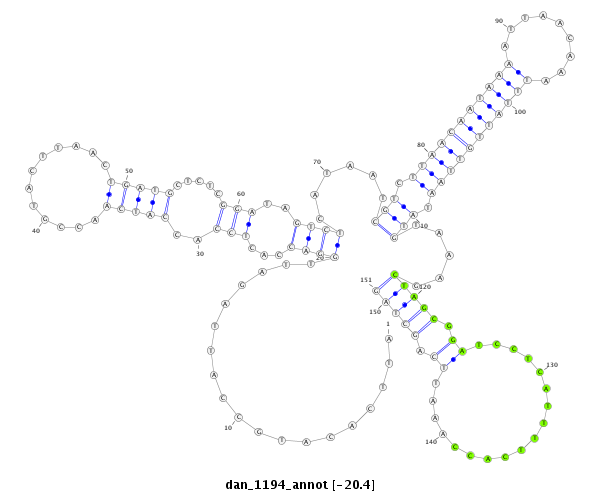
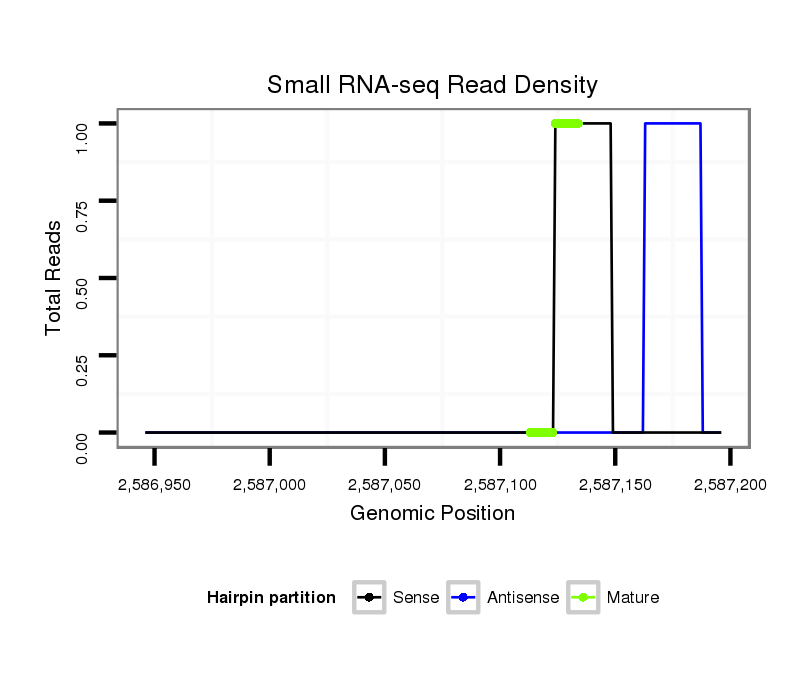
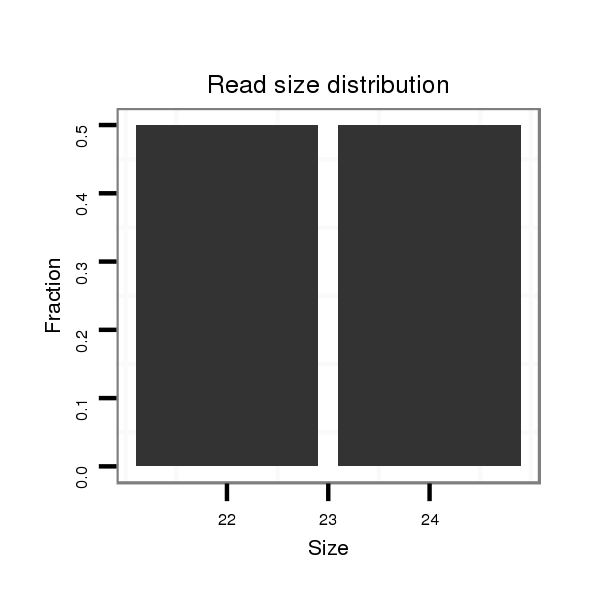
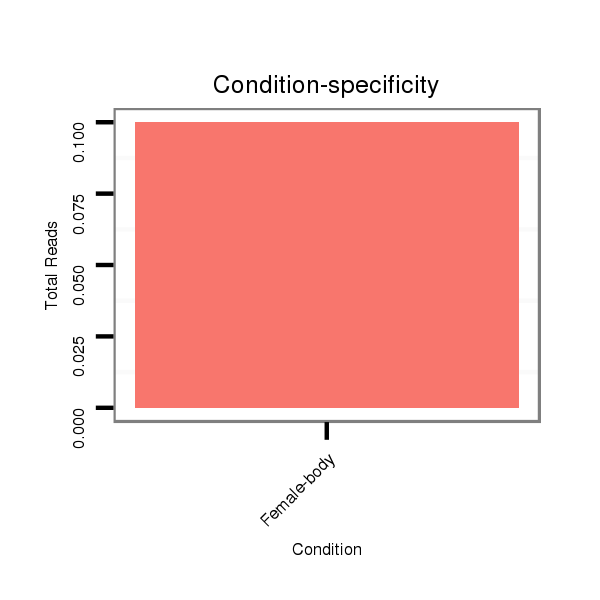
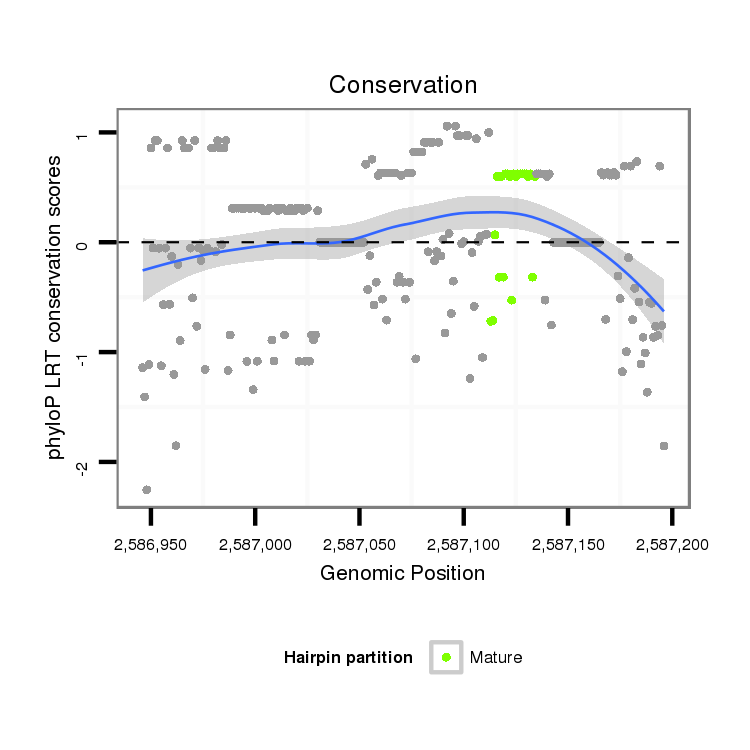
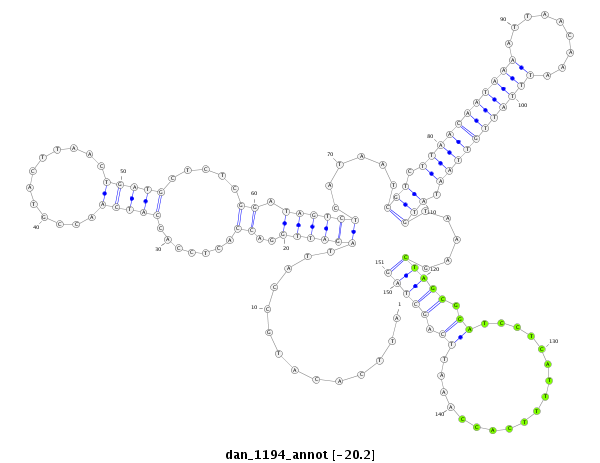
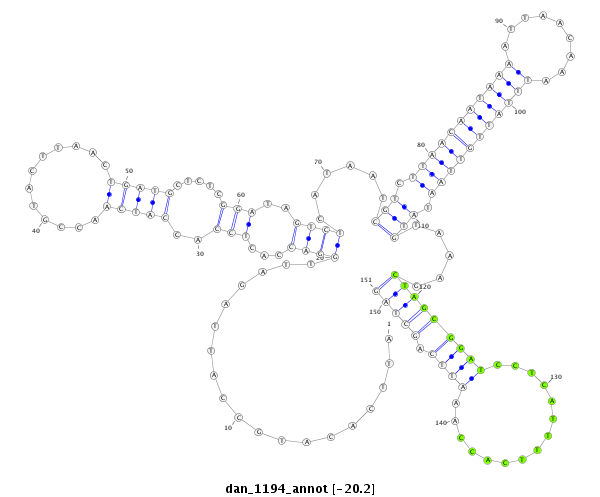
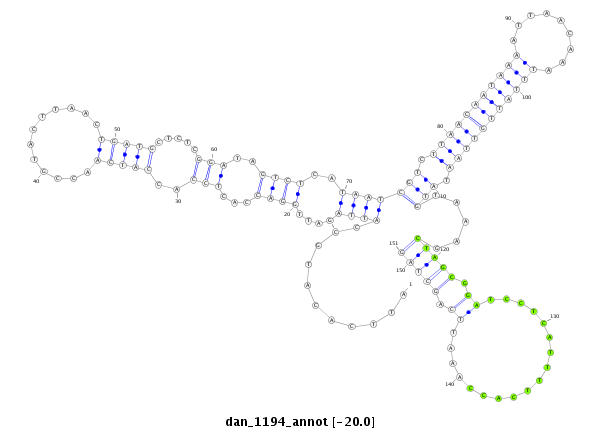
ID:dan_1194 |
Coordinate:scaffold_13082:2586996-2587146 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.2 | -20.2 | -20.0 |
 |
 |
 |
CDS [Dana\GF20563-cds]; exon [dana_GLEANR_3356:4]; intron [Dana\GF20563-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGCGGATTTATTGAAGCGAACGGATATTTCCACGGCACGCAAGTTTATATATTCACATGCCATTAGATTGGACCACTCCACCATCAACCGTACTTAACTGATGCTCTCGGATAGTCTCATAATCGTCTTAACAATAAAATTAACAAATTTATTGTTAATATGTAAAGCTAGCGGATCCTCATTTTCACCAAATTCAGCTAGCCTCAGACTGAGCTGGTGAAGGTGGAGTCCCATCGCAGTTCCGTTTCCAA **************************************************...................((((...(((..(((((............))))).....)))..))))......(((.(((((((((((.........))))))))))).))).....(((((.((..................)).)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V039 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TCATTTTCACCAAATTCAGCTAGCC................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................TAAATTGGACCACCCCACC......................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................ACCAACCGTACTTAACGG....................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................GCTGTTGAAGGTGGATTTCCA.................. | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 |
| ..........TTGAAGCGAACGGATATTTCCACGGCAC..................................................................................................................................................................................................................... | 28 | 0 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| ..........TTGAAGCGAACGGATATTTCCAAGG........................................................................................................................................................................................................................ | 25 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ................................................................AGATGGGACCAAGCCACCA........................................................................................................................................................................ | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AAGACTGGGGTGGTGAAGG............................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................CTAGCGGATCCTCATTTTCACC.............................................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................ATGTACACCAGATTCAGCT.................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................AGCTAGCGGATCCTCATTTTCACC.............................................................. | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
TCGCCTAAATAACTTCGCTTGCCTATAAAGGTGCCGTGCGTTCAAATATATAAGTGTACGGTAATCTAACCTGGTGAGGTGGTAGTTGGCATGAATTGACTACGAGAGCCTATCAGAGTATTAGCAGAATTGTTATTTTAATTGTTTAAATAACAATTATACATTTCGATCGCCTAGGAGTAAAAGTGGTTTAAGTCGATCGGAGTCTGACTCGACCACTTCCACCTCAGGGTAGCGTCAAGGCAAAGGTT
**************************************************...................((((...(((..(((((............))))).....)))..))))......(((.(((((((((((.........))))))))))).))).....(((((.((..................)).)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V055 head |
V105 male body |
V039 embryo |
V106 head |
M058 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................GAATTGACTACCAGACCCGA........................................................................................................................................... | 20 | 3 | 10 | 1.00 | 10 | 4 | 2 | 1 | 0 | 3 | 0 |
| .........................................................................................................................................................................................................................ACTTCCACCTCAGGGTAGCGTCAAG......... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................GAATTGACTACCAGACCC............................................................................................................................................. | 18 | 2 | 4 | 1.00 | 4 | 0 | 4 | 0 | 0 | 0 | 0 |
| ............................................................................................GAATTGACTACCAGACCCG............................................................................................................................................ | 19 | 3 | 20 | 0.55 | 11 | 4 | 0 | 0 | 0 | 7 | 0 |
| .......................................................................TGGGGAGTTGGTAGTTGGCAGG.............................................................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................AACATGGTGAGTTGGTAGT..................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GGGTATCGTCAAGGGGAAGGT. | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................ACATGGTGAGTTGGTAGT..................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................ATTTCGATCGCCTAGGAGTAAAAGTG............................................................... | 26 | 0 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GAGGTGGGAGTTGGCATA.............................................................................................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................CGATCGCCTAGGAGTAAAAGTG............................................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| ...................................................................................................................................................................TTTCGATCGCCTAGGAGTAAAAGTG............................................................... | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................TTCGATCGCCTAGGAGTAAAAGTGGT............................................................. | 26 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................ATTTCGATCGCCTAGGAGTAAAAGT................................................................ | 25 | 0 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................AATTTCGATCGCCTAGGAGTAAAAGTG............................................................... | 27 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TACATTTCGATCGCCTAGGAGTAAAA.................................................................. | 26 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................CATTTCGATCGCCTAGGAGTAAAA.................................................................. | 24 | 0 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................TTCGATCGCCTAGGAGTAAAAG................................................................. | 22 | 0 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................TCGATCGCCTAGGAGTAAAAGGG............................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................TCGATCGCCTAGGAGTAAATCGG............................................................... | 23 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13082:2586946-2587196 + | dan_1194 | AGCGGATTTATTGAAGCGAACGGATATTTCCACGGCACGCAA--------------------------------------------GTTTATATATTCACATGCCATTAGATTGGACCACTCCACCATCAACCGTACTTAACTGATGCTCTCGGATAGTCTCATAATCGTCTTAACAATAAAATTAACAAATTTATTGTTAA------T-ATGTAAA-GCTAGCGGATCCTCATTTTCACCAAATTCAGCTAGCCTCAGACTGAGCTGGTGAAGGTGGAGTCCCATCGCA-GTTCCGTTTCCAA |
| droBip1 | scf7180000395259:4562-4742 + | AGTGGATTTGTTGAAAGGGACGGAGATTTTTATGGCACGCAGTCAAAGAATAAATTTAAAGTGTAATAAAACTGCTGTCAGGCTATATTTATATCTTGAAATGCCAAGAGATCGGACCAATCAAATTCC--------------------------------------------------AAAATTAACTTATTTA------G------T-TTCATAAAGTCA------------------------------------------------------------GCCTCACAGAAGAAACGAACTT | |
| droKik1 | scf7180000302230:166076-166079 - | CCA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A | |
| droFic1 | scf7180000453221:58657-58697 - | ATCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGGA-GTTGGTGAATCATCATTTTCATCAAATGCAA------------------------------------------------------C | |
| droRho1 | scf7180000774099:791-831 + | AGGTGATTTAATGG--ATAACGGATAGTACCACGGAACACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G | |
| droBia1 | scf7180000302069:1423285-1423324 + | AGAC---------------------------------------------------------------------------------------------------------------------------------------------------CGAAC--------------------------------------------------------------------------------------------------------------------AGCTGGAGCAGCACCAGA-GCGAGGCGACCAA | |
| droTak1 | scf7180000410478:1222-1276 - | CTCC---------------------------------------------------------------------------------------------------------------------------------------------------CAAATTGTATGATAAATGAATAAATAATAAAATAAATAAATAAATTATTA------------------------------------------------------------------------------------------------------A | |
| droEug1 | scf7180000409128:4544-4621 - | GGAGGGTTATCCGTTCTGGACGGGTACCTCATCGGAACACAT--------------------------------------------------------------------------------------------------------------------------------------------------ATATCAATTGCTATTGCTAGACCTCTTACCGCCA--------------------------T------------------------------------------------------C | |
| droSec2 | scaffold_933:546-588 - | TTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAATAAGATTTACAAACTTATTGTTAG------T-CTCTTAA-GTTA---------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/18/2015 at 12:47 AM