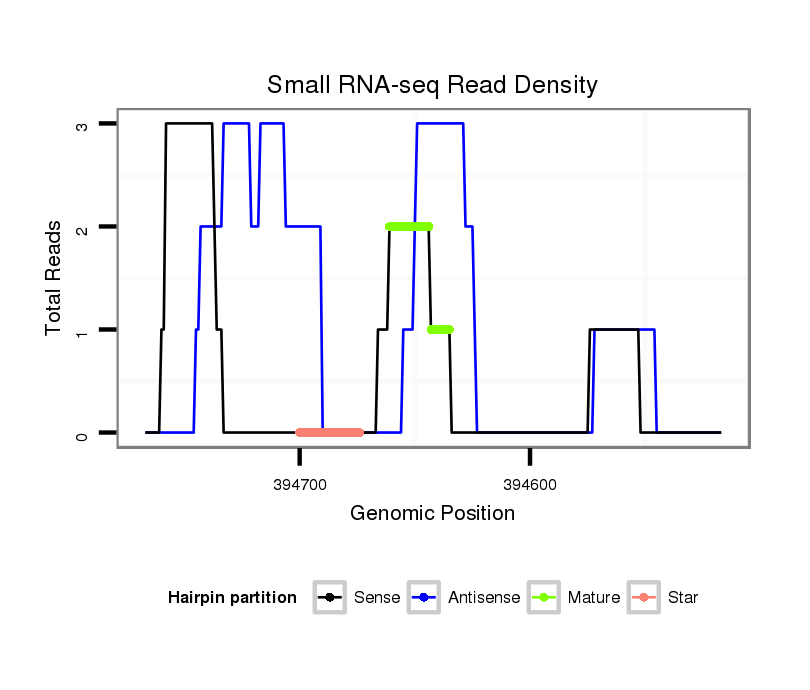
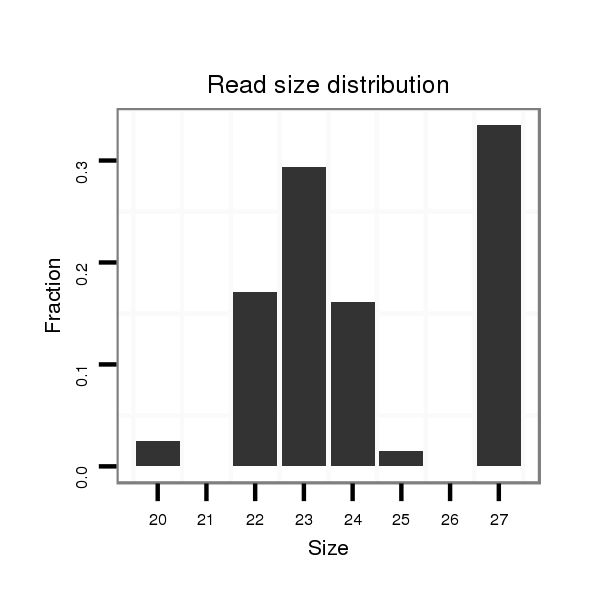
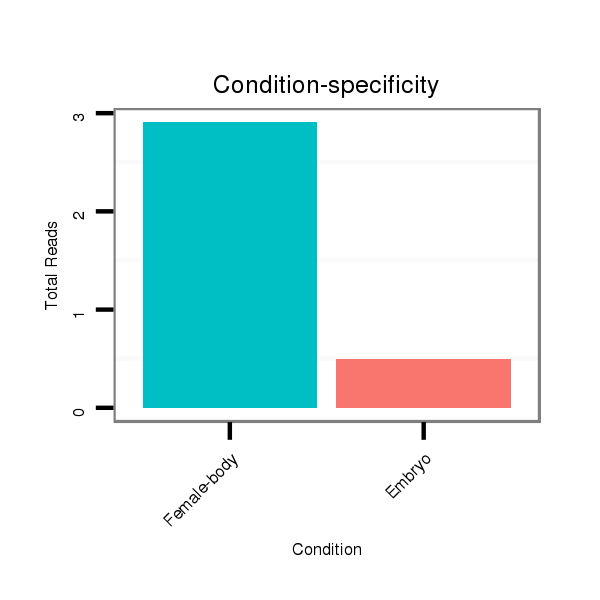
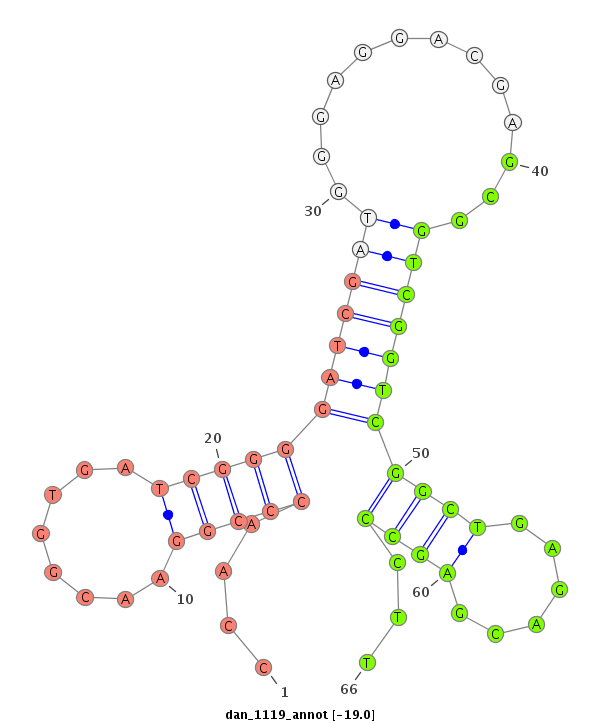
| TTGGCACCAC-------C-----AGACCCTGCAC---------ATCACGTGGGCCTCGGG------GCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------GGG-------------------------------------------------- | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| ....CACCAC-------C-----AGACCCTGCAC---------ATCAC................................................................................................................................................................................................................................................ | 23 | 13 | 5.00 | 65 | 0 | 64 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------ATCACG............................................................................................................................................................................................................................................... | 24 | 13 | 3.92 | 51 | 0 | 50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................AC---------ATCACGTGGGCCTCGGG------GCG........................................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TGCAC---------ATCACGTGGGCCTCGGG------GCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------..................................................... | 26 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------ATCA................................................................................................................................................................................................................................................. | 22 | 13 | 1.54 | 20 | 0 | 18 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------ATCACGT.............................................................................................................................................................................................................................................. | 25 | 13 | 1.38 | 18 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------ATC.................................................................................................................................................................................................................................................. | 21 | 13 | 1.23 | 16 | 0 | 15 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TGCAC---------ATCACGTGGGCCTCGGG------GCG........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GCAC---------ATCACGTGGGCCTCGGG------GCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------GG................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................C---------ATCACGTGGGCCTCGGG------GCGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------G.................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------AT................................................................................................................................................................................................................................................... | 20 | 13 | 0.23 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ACCAC-------C-----AGACCCTGCAC---------ATCAC................................................................................................................................................................................................................................................ | 22 | 13 | 0.23 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------A.................................................................................................................................................................................................................................................... | 19 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ACCAC-------C-----AGACCCTGCAC---------ATCACGT.............................................................................................................................................................................................................................................. | 24 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------ATCACGTGG............................................................................................................................................................................................................................................ | 27 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ACCAC-------C-----AGACCCTGCAC---------ATCA................................................................................................................................................................................................................................................. | 21 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ACCCTGCAC---------ATCACGTGGGCCTCG...................................................................................................................................................................................................................................... | 24 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CCTGCAC---------ATCACGTGGGCCT........................................................................................................................................................................................................................................ | 20 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................AC---------ATCACGTGGGCCTCGGG------GC............................................................................................................................................................................................................................ | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ACCCTGCAC---------ATCACGTGGGCCTCGGG------.............................................................................................................................................................................................................................. | 26 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TGCAC---------ATCACGTGGGCCTCGGG------.............................................................................................................................................................................................................................. | 22 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .TGGCACCAC-------C-----AGACCCTGCAC---------ATCACG............................................................................................................................................................................................................................................... | 27 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CCAC-------C-----AGACCCTGCAC---------ATCAC................................................................................................................................................................................................................................................ | 21 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...GCACCAC-------C-----AGACCCTGCAC---------ATCACG............................................................................................................................................................................................................................................... | 25 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ACCAC-------C-----AGACCCTGCAC---------ATCACG............................................................................................................................................................................................................................................... | 23 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ACCAC-------C-----AGACCCTGCAC---------ATCACGTGGGC.......................................................................................................................................................................................................................................... | 28 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ACCAC-------C-----AGACCCTGCAC---------A.................................................................................................................................................................................................................................................... | 18 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CACCAC-------C-----AGACCCTGCAC---------ATCACGTG............................................................................................................................................................................................................................................. | 26 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CAC-------C-----AGACCCTGCAC---------ATCACGTGG............................................................................................................................................................................................................................................ | 24 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................AGACCCTGCAC---------ATCACGTGGG........................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CTGCAC---------ATCACGTGGGCCTC....................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CCCTGCAC---------ATCACGTGGGCCTC....................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................AC---------ATCACGTGGGCCTCGGG------G............................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CCTGCAC---------ATCACGTGGGCCTCGG..................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................C-----AGACCCTGCAC---------ATCACGTGGG........................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CCCTGCAC---------ATCACGTGGGCC......................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CCTGCAC---------ATCACGTGGGCCTC....................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ACCCTGCAC---------ATCACGTGGGCCT........................................................................................................................................................................................................................................ | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CAC---------ATCACGTGGGCCTCGGG------GC............................................................................................................................................................................................................................ | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................ATCACGTGGGCCTCGGG------GC............................................................................................................................................................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ACCCTGCAC---------ATCACGTGGG........................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CCTGCAC---------ATCACGTGGGCCC........................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TGCAC---------ATCACGTGGGCCTCGGG------GC............................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ACCCTGCAC---------ATCACGTGGGCCTCGG..................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |