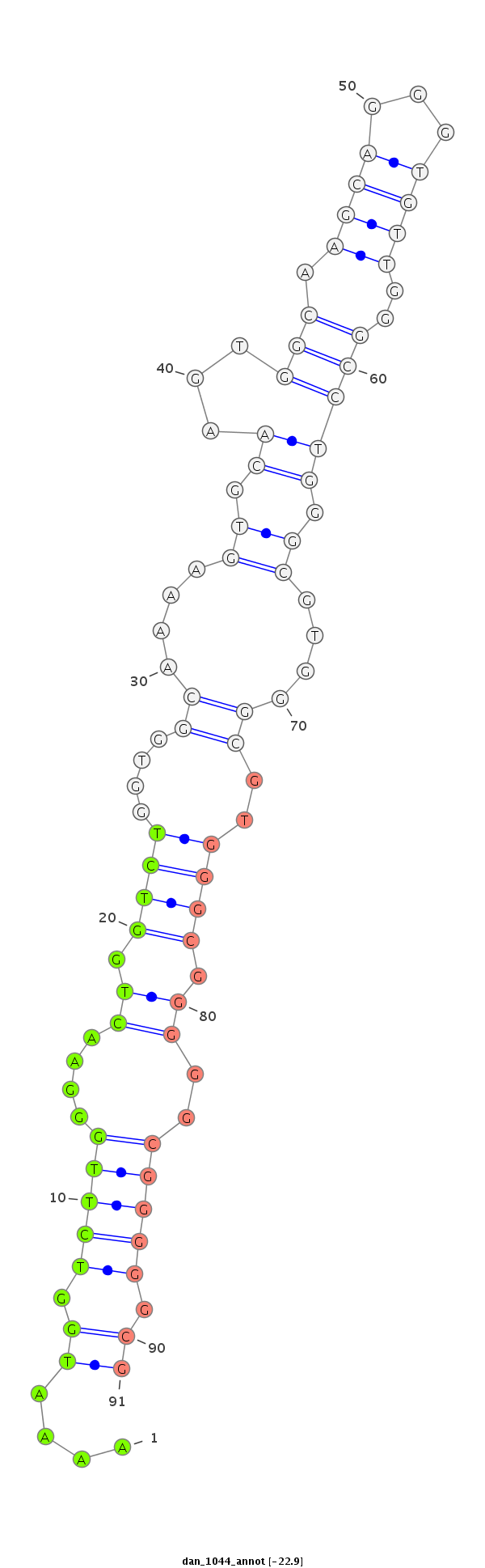
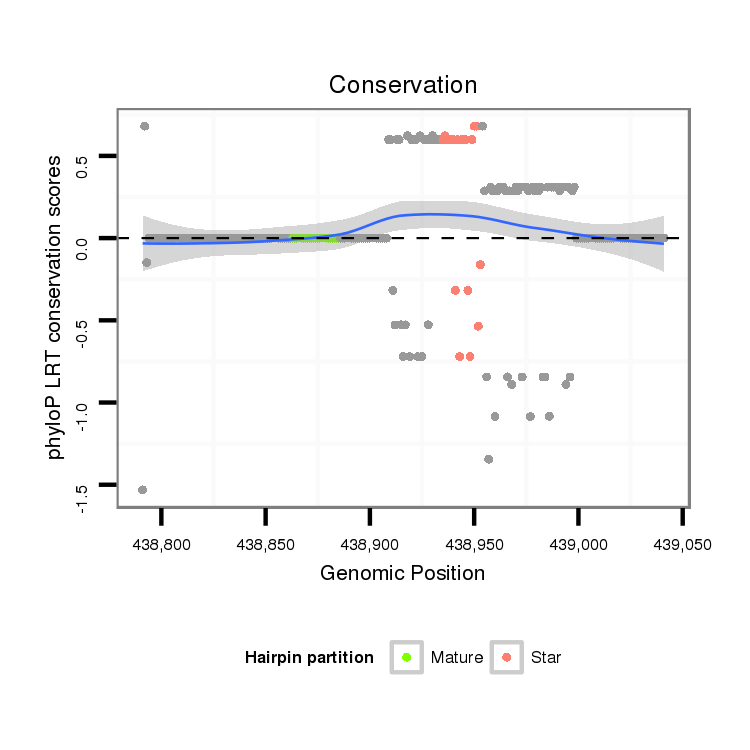
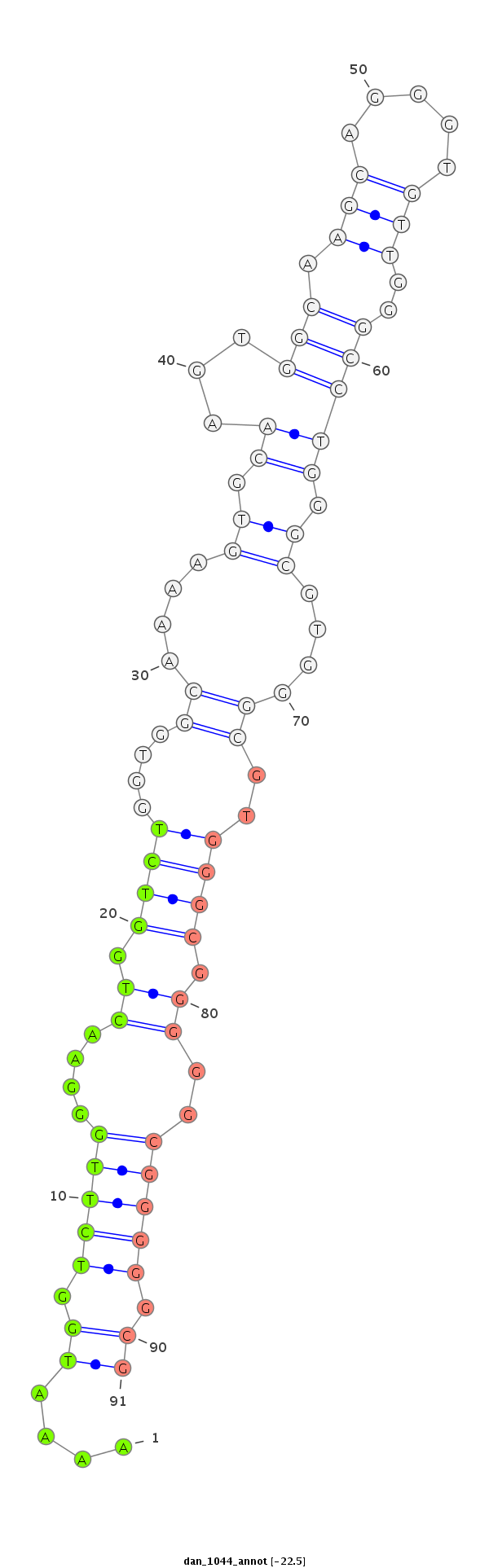
ID:dan_1044 |
Coordinate:scaffold_12948:438841-438991 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
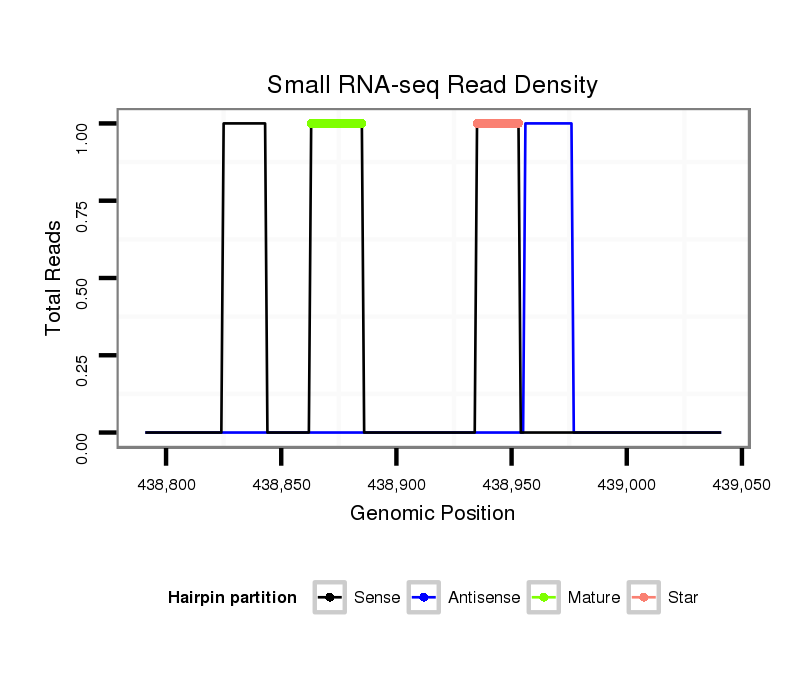
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.5 | -22.5 | -22.3 |
 |
 |
 |
exon [dana_GLEANR_5104:1]; CDS [Dana\GF21594-cds]; intron [Dana\GF21594-in]
| Name | Class | Family | Strand |
| (CGGGGG)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CGACCCGCATAGTATTACCGGCCTGGTATCAGTTCCGGAGCTGCGCAAGAGTGAGTCGCCTCTAAATTGCACAAAATGGTCTTGGGAACTGGTCTGGTGGCAAAAGTGCAAGTGGCAAGCAGGGTGTTGGGCCTGGGCGTGGGCGTGGGCGGGGGCGGGGGCGGGTGTGTCATTGGCAGACTGAAGCAGACTCGAAATAACAATAGCACCACTTCCCACAGCTTTTGATACTTTCTTGAAGGCGGAAGAGT ************************************************************************....((.(((((....((.((((....((....((.((...(((.((((...))))..))))).))....))..)))).))..))))).))**************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V039 embryo |
V055 head |
V105 male body |
V106 head |
M058 embryo |
M044 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................AGTTTTTGATACTTTCTTGA............ | 20 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTTTTGATACTTTCTTGA............ | 19 | 1 | 2 | 2.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................AAAATGGTCTTGGGAACTGGTCT............................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................CCGGAGCTGCGCAAGAGTG...................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................GTGGGCGGGGGCGGGGGCG........................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................GCTTTTGATACTTTCTTCA............ | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTTTTGATACTTTCTTGAA........... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GCTTTTGATACTTTCGTGA............ | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................CGGGGGCGGGGGTAGGTGTGT................................................................................. | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................ATTGGTGGACTGAAGCAGA............................................................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GCTTTCGATACTTTCTGGA............ | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................GGCCGGGGTGGCATTGGCAGAC...................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................GAGGGGCGGGGGCGGGTGTG.................................................................................. | 20 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTTTTGATACTTTCTCGA............ | 19 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TTGGAAGACTGAAGCGGA............................................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................GCGGGTGTGGAATTGGTAGAC...................................................................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTTTTGATACTTTCTAGA............ | 19 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................CGACAGAACAATAGCACTAC....................................... | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GGCGCGGCGGCGGGTGTG.................................................................................. | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................................................AGGTTTTGATACTTTCCCGA............ | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........ATAGTATTACCGGAACGGT................................................................................................................................................................................................................................ | 19 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTTTTAATACTTTCTTGA............ | 19 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................GTGCCTGGGCGTTCGCGTG........................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................GTGGGCGGGGGGAGGGGCG........................................................................................ | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................TGGGGCCGGGCGCGGGCGTG........................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GCTGGGCGTATCATAATGGCCGGACCATAGTCAAGGCCTCGACGCGTTCTCACTCAGCGGAGATTTAACGTGTTTTACCAGAACCCTTGACCAGACCACCGTTTTCACGTTCACCGTTCGTCCCACAACCCGGACCCGCACCCGCACCCGCCCCCGCCCCCGCCCACACAGTAACCGTCTGACTTCGTCTGAGCTTTATTGTTATCGTGGTGAAGGGTGTCGAAAACTATGAAAGAACTTCCGCCTTCTCA
****************************************************************************************....((.(((((....((.((((....((....((.((...(((.((((...))))..))))).))....))..)))).))..))))).))************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
M058 embryo |
V105 male body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................ACACAGTAACCGTCTGACTTC................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................ACCGTCTTAGTTCGTCTGA........................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TGATATGGTGGAGAAGGGTGT............................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................CCACAGCGCGAACCCGCACCCG........................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................CCGCCCCCCCCCACACA................................................................................. | 17 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................AGGGTGTCCGATACTATGA................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................TGCAGGGTGTCGACAACT....................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................GCCGTCCGAGTTCGTCTGA........................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................TAACCGTCATACTTCGTGT............................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_12948:438791-439041 + | dan_1044 | CGACCCGCATAGTATTACCGGCCTGGTATCAGTTCCGGAGCTGCGCAAGAGTGAGTCGCCTCTAAATTGCACAAAATGGTCTTGGGAACTGGTCTGGTGGCAAAAGTGCAAGTGGCAAGCAGGGTGTTGGGCCTGGGCGTGGGCGTGGGCGGGGGCGGGGGCGGGTGTGTCATTGGCAGACTGAAGCAGACTCGAAATAACAATAGCACCACTTCCCACAGCTTTTGATACTTTCTTGAAGGCGGAAGAGT |
| droBip1 | scf7180000395902:38113-38164 - | GGA------------------------------------------------------------------------------------------------------------------------------------------------------------GGTGGGCCTGGCATTGACTGACTAAAGAAGACTTAACATAACAAAAACA------------------------------------------- | |
| droFic1 | scf7180000453906:839118-839166 + | TGG-------------------------------------------------------------------------------------------------------------------GCGTGGGCGTCGGCGTCGGAGTGGGCGTGGGCAGCGGCACGGGCAG--------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
Generated: 05/18/2015 at 12:03 AM