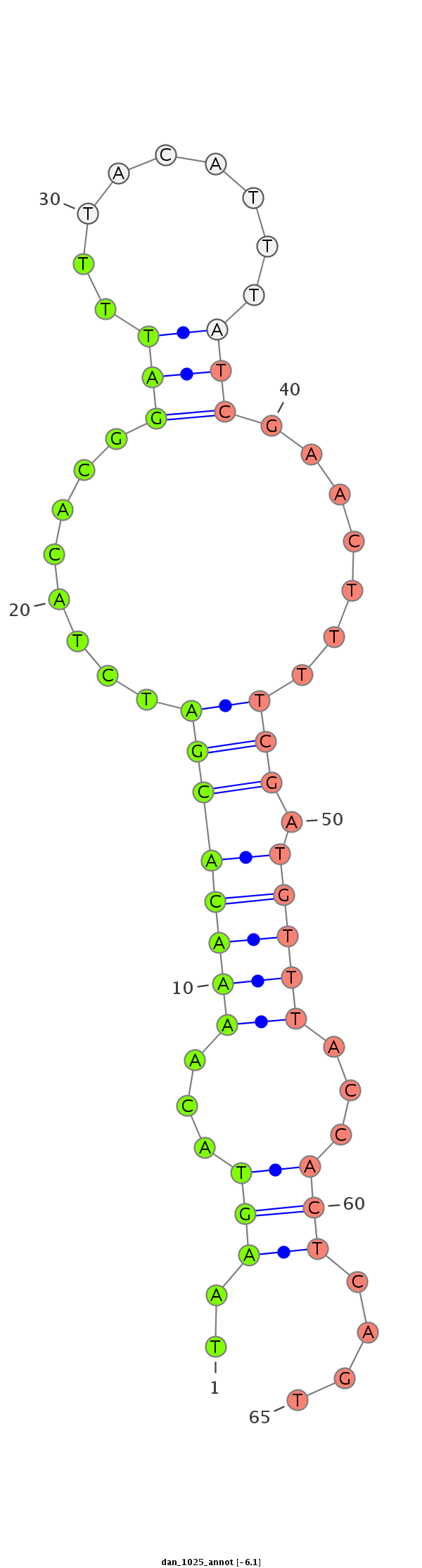
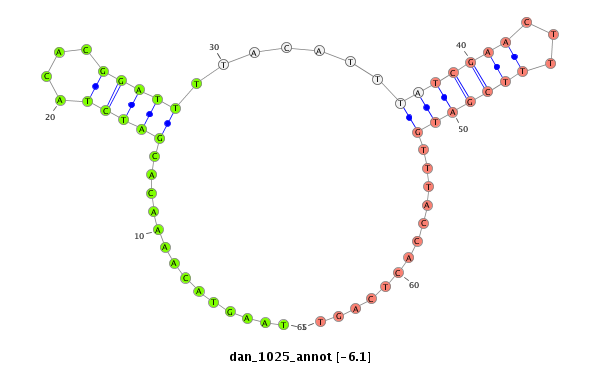
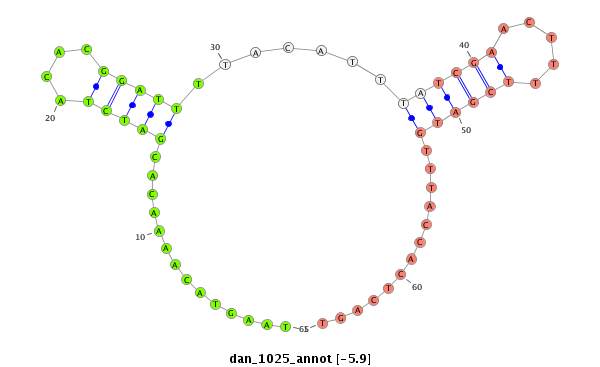
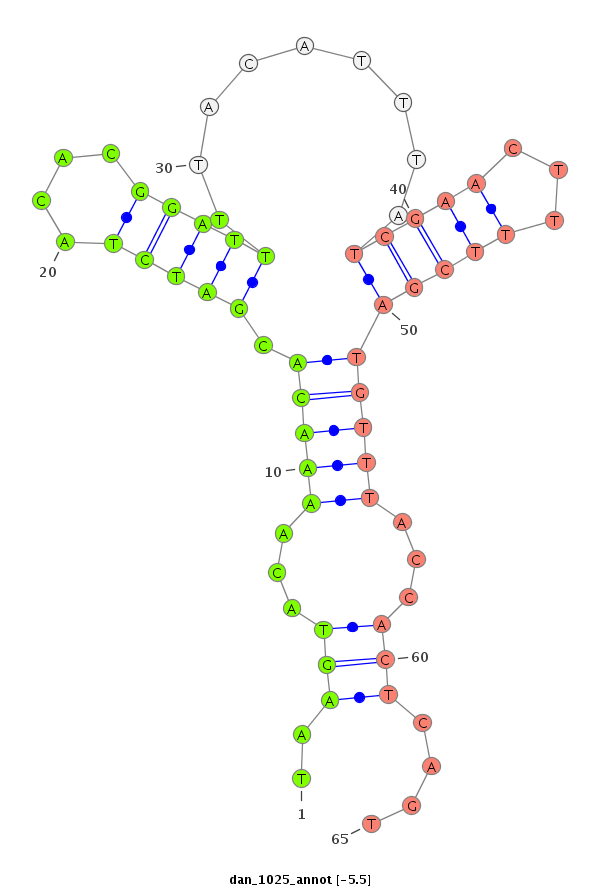
| ...................................................TAAGTACAAAACACGATCTACACGGATTT........................................................................................................................................................................... |
29 |
0 |
1 |
7.00 |
7 |
7 |
0 |
0 |
0 |
0 |
| ...................TACAACCAATCGACACAACCAGCAGCAC............................................................................................................................................................................................................ |
28 |
0 |
1 |
6.00 |
6 |
5 |
0 |
1 |
0 |
0 |
| ...................................................TAAGTACAAAACACGATCTACACGGA.............................................................................................................................................................................. |
26 |
0 |
1 |
5.00 |
5 |
5 |
0 |
0 |
0 |
0 |
| ......................AACCAATCGACACAACCAGCAGCACGA.......................................................................................................................................................................................................... |
27 |
0 |
1 |
5.00 |
5 |
5 |
0 |
0 |
0 |
0 |
| ..........TACAACATGTACAACCAATCGACACAAC..................................................................................................................................................................................................................... |
28 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
| .........................................................................................CAAACGTTTCGATGTTTACC.............................................................................................................................................. |
20 |
2 |
1 |
3.00 |
3 |
0 |
3 |
0 |
0 |
0 |
| .................TGTACAACCAATCGACACAACCAGCAGC.............................................................................................................................................................................................................. |
28 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| .........................CAATCGACACAACCAGCAGCACGATG........................................................................................................................................................................................................ |
26 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| ........................................................................................TCGAACTTTTCGATGTTTACCACTCAGT....................................................................................................................................... |
28 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ...................................................................................................GATGTTTACCACTCAGTGACTTTAGT.............................................................................................................................. |
26 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ...................................................................................................GATGTTTACCACTCAGTGACTTTAGTT............................................................................................................................. |
27 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..............ACATGTACAACCAATCGACACAACC.................................................................................................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............................TCGACACAACCAGCAGCACGATGTAAG.................................................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................TTTACTATGACGATCTTTATTTCTTTGTC........................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............................................................CACGATCTACACGGATTTTACATTTATC................................................................................................................................................................. |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................ACCAGCAGCACGATGTAAGTACAAAACAC.......................................................................................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................TTTACCACTCAGTGACTTTAGTTTCTTA........................................................................................................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................................................AACTTTTCGATGTTTACCACTCAGTGAC.................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................ACAACCAATCGACACAACCAGCAGCAC............................................................................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................CAATCGACCCAACCAGCAGCACGATG........................................................................................................................................................................................................ |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................GCAGCACGATGTAAGTACAAAACACGATC...................................................................................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................CAGCAGCACGATGTAAGTACAAAACAC.......................................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................TAAGCTGCAAAATTATTGCTTTCCCC....................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................TAAGCTGCAAAATTATTGCTTTCCCCT...................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................ACCACTCAGTGACTTTAGTTTCTTAAAC..................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............CATGTACAACCAATCGACACAACCAGCA................................................................................................................................................................................................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................AAATTATTGCTTTCCCCTTCCAAGCT.............................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................TCGATGTTTACCACTCAGTGACTTTAGT.............................................................................................................................. |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................ATGTACAACCAAACGGCACCAC..................................................................................................................................................................................................................... |
22 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .............................................................................................................................................................................................TATTATTTTACTATCACGATCTTTATT................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................TACAACCAATCGACACAACCAGCAGCA............................................................................................................................................................................................................. |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................TTCGATGTCTACCACTCAGTGACTTTAGT.............................................................................................................................. |
29 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................................TTTACCACTCAGTGACTTTAGTTTCTT......................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................AAGTACAAAACACGATCTACACGGATT............................................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................GCAGCACGATGTAAGTACAAAACACGA........................................................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................AATTATTGCTGTCCCCTCC.................................................................................... |
19 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................TACACGGATTTTACATTTATCGAACTTT.......................................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................TTTCGATGTTTACCACTCAGTGACTT.................................................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................CACGGATTTTACATTTATCGAACTTT.......................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........TACAACATGTACAACCAATCGACACAACC.................................................................................................................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............................................................................................CTTTTCGATGTTTACCACTCAGTGACTT.................................................................................................................................. |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................TGCAAAATTATTGCTTTCCCCTTCCAAGC............................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................TATTTTACTATCACGATCTTTATTTCTT............................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................CCAGCAGCACGATGTAAGTACAAAACAC.......................................................................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................TGTTTACCACTCAGTGACTTTAGTTTCT.......................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................AACCAGCAGCACGATGTAAGTACAAAA............................................................................................................................................................................................. |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............CAACATGTACAACCAATCGACACAAC..................................................................................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................CCAATCGACACAACCAGCAGCACGATG........................................................................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................TGTAAGTACAAAACACGATCTACACGG............................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................TACAAAACACGATCTACACGGATTT........................................................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....TCGATTACAACATGTACAACCAATCGAC.......................................................................................................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................TAAGTACAAAACACGATCTACACGGATT............................................................................................................................................................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................ACCAATCGACACAACCAGCAGCACGATG........................................................................................................................................................................................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................ACCAATCGACACAACCAGCAGCACGA.......................................................................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TCCCCTTCCAAGCTTATTTAATCCTAAC................................................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................................AACTCTATTATTTTACTATCACGATCT........................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................TACCACTCAGTGACTTTAGTTTCTTAAAC..................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............................................ACGATGTAAGTACAAAACACGATC...................................................................................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................AGTACAAAACACGATCTACACGGATTT........................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................AACCAATCGACACAACCAGCAGCACGATG........................................................................................................................................................................................................ |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........TACAACATGTACAACCAATCGACACA....................................................................................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............................CGACACAACCAGCAGCACGATGTAAGT................................................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................CAACCAGCAGCACGATGTAAGTACAAAA............................................................................................................................................................................................. |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............AACATGTACAACCAATCGACACAACCA................................................................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............................GACACAACCAGCAGCACGATGTAAGTACA................................................................................................................................................................................................ |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..............................................................................................TTTTCGATGTTTACCACTCAGTGACTT.................................................................................................................................. |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................................................................................TATGTGAAATTCTTTTAC. |
18 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
| ....................................................................................................................................................AATTATTGCTGTCCCCCACC................................................................................... |
20 |
3 |
6 |
0.33 |
2 |
0 |
2 |
0 |
0 |
0 |
| ...................TACAACCATTCGAAACAACT.................................................................................................................................................................................................................... |
20 |
3 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
1 |
| ....................................................................................................................................................AATTATTGCTGTCCCTTACCA.................................................................................. |
21 |
3 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..............................................TGATGGAAGTACGAAACACG......................................................................................................................................................................................... |
20 |
3 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
1 |
| ..........................AAGCGACACAACCCGCACC.............................................................................................................................................................................................................. |
19 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
| .........................................................................................................................TAGTTTCTTCAACTTCTTATGCT........................................................................................................... |
23 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |
| ....................................................................................................................................................AATTATTGCTGTCCCCCAC.................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................AGTTTCTTAATCTGATTA............................................................................................................... |
18 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |