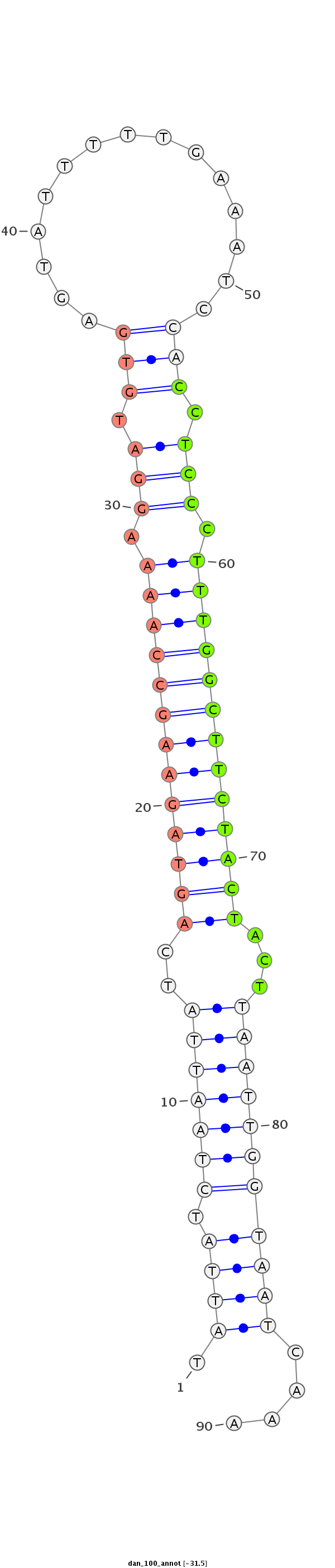
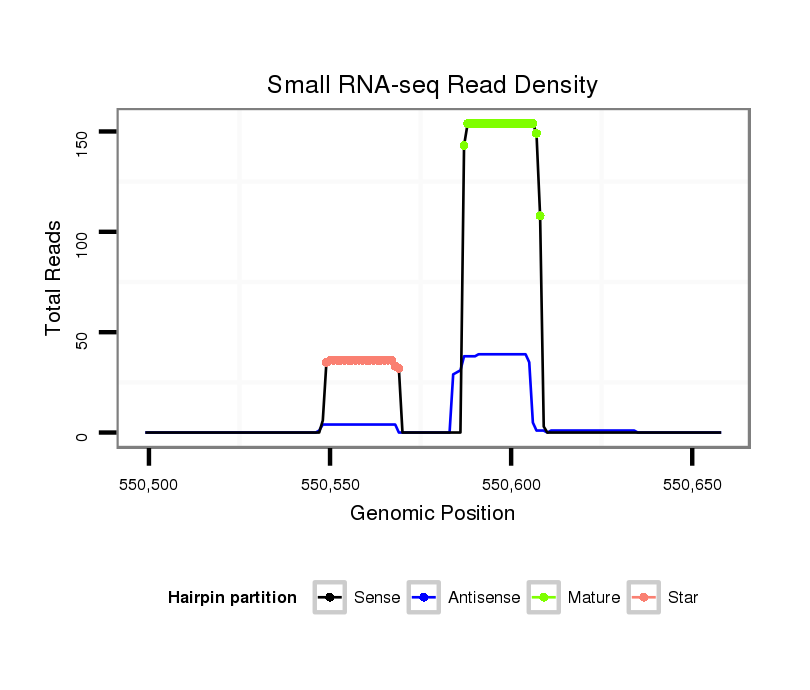
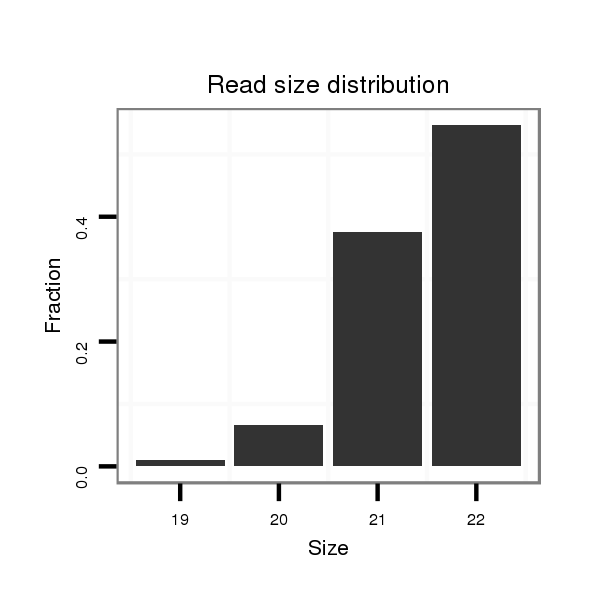
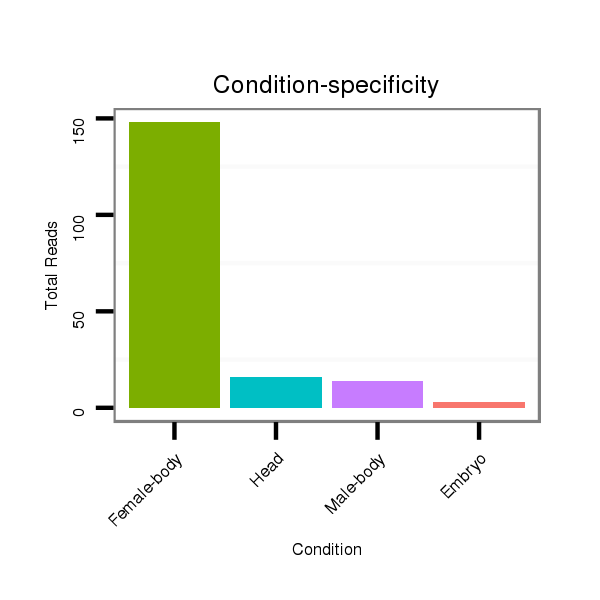

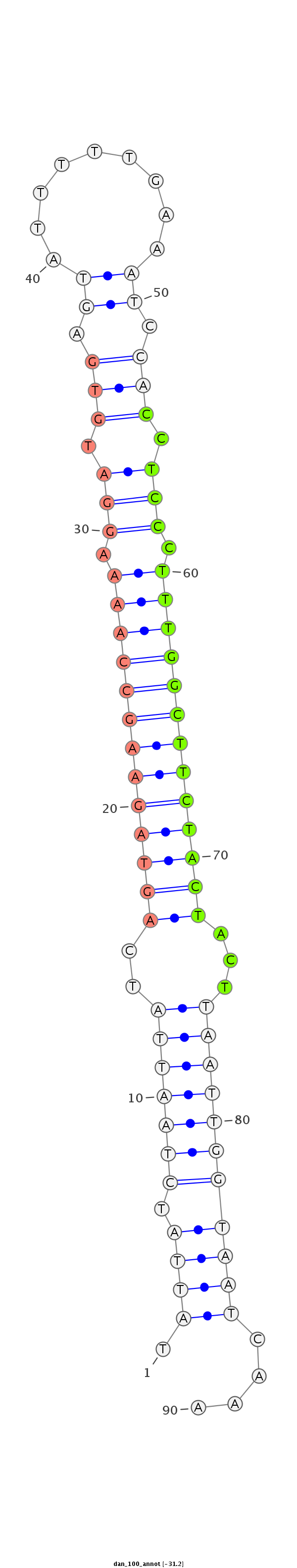
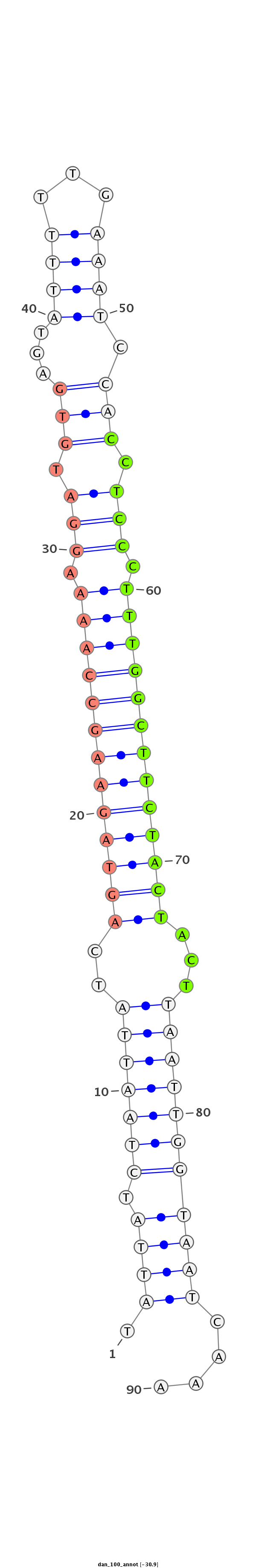
ID:dan_100 |
Coordinate:scaffold_12903:550549-550608 + |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:antisense_to_intron |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.2 | -31.2 | -30.9 |
 |
 |
 |
Antisense to intron [Dana\GF15947-in]
No Repeatable elements found
|
AAAAAATTTTTAAAAAGTGATAATAATTCTTGGAGTATTATCTAATTATCAGTAGAAGCCAAAAGGATGTGAGTATTTTTGAAATCCACCTCCCTTTGGCTTCTACTACTTAATTGGTAATCAAAACTGCTTCAAAAACAGTCTAAAACTAACTGAAAAG
***********************************.((((.(((((((..(((((((((((((.(((.(((...............))).))).)))))))))))))...)))))))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V105 male body |
V106 head |
V055 head |
M058 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................CCTCCCTTTGGCTTCTACTACT.................................................. | 22 | 0 | 1 | 99.00 | 99 | 96 | 1 | 0 | 0 | 2 |
| ........................................................................................CCTCCCTTTGGCTTCTACTAC................................................... | 21 | 0 | 1 | 36.00 | 36 | 32 | 1 | 2 | 1 | 0 |
| ..................................................AGTAGAAGCCAAAAGGATGTG......................................................................................... | 21 | 0 | 1 | 26.00 | 26 | 4 | 11 | 9 | 1 | 1 |
| .........................................................................................CTCCCTTTGGCTTCTACTACT.................................................. | 21 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 |
| .........................................................................................CTCCCTTTGGCTTCTACTAC................................................... | 20 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 |
| ........................................................................................CCTCCCTTTGGCTTCTACTA.................................................... | 20 | 0 | 1 | 5.00 | 5 | 4 | 0 | 1 | 0 | 0 |
| .................................................CAGTAGAAGCCAAAAGGATGTG......................................................................................... | 22 | 0 | 1 | 5.00 | 5 | 0 | 4 | 1 | 0 | 0 |
| ........................................................................................CCTCCCTTTGGCTTCTACTACTT................................................. | 23 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ........................................................................................CCTCCCTTGGGCTTCTACTACT.................................................. | 22 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..................................................AGTAGAAGCCAAAAGGATG........................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 |
| ........................................................................................CCTCCCTTTGGCTTCTACTACTA................................................. | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ........................................................................................CCTCCCTTGGGCTTCTACTAC................................................... | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................AGTAGAAGCCAAAAGGATGTGTT....................................................................................... | 23 | 2 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 |
| ..................................................AGTAGAAGCCAAAAGGATGTC......................................................................................... | 21 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 |
| ..................................................AGTAGAAGCCAAAAGAATGTG......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................CCTCCCTTCGGCTTCTACTA.................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................GTAGAAGCCAAAAGGATGTG......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTCCCTTTGGCTTCTACTAC................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................CCTCCCTTTGGCTTCTACTACC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................AGTAGAAGCCAAAAGGATGTA......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GGTAGAAGCCAAAAGGATGTG......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................CCTCCCTTTGGCTTCAACTACT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................CCTCCCTTAGGCTTCTACTACT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................AGTAGAAGCCAAAAGGATGTGTTT...................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................CAGTAGAAGCCAAAAGGATG........................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................ACTCCCTTTGGCTTCTACTAC................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TCTCCCTTTGGCTTCTACTACT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................AGTAGAAGGCAAAAGGATGTG......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................AGTAGAAGCCAAAAGGATGT.......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................AAAAACAGTCAAAAGCTAAC....... | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TTTTTTAAAAATTTTTCACTATTATTAAGAACCTCATAATAGATTAATAGTCATCTTCGGTTTTCCTACACTCATAAAAACTTTAGGTGGAGGGAAACCGAAGATGATGAATTAACCATTAGTTTTGACGAAGTTTTTGTCAGATTTTGATTGACTTTTC
***********************************.((((.(((((((..(((((((((((((.(((.(((...............))).))).)))))))))))))...)))))))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
V105 male body |
M044 female body |
M058 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................GGTGGAGGGAAACCGAAGATGA..................................................... | 22 | 0 | 1 | 24.00 | 24 | 11 | 9 | 4 | 0 | 0 |
| .....................................................................................GGTGGAGGGAAACCGAAGATG...................................................... | 21 | 0 | 1 | 4.00 | 4 | 1 | 1 | 2 | 0 | 0 |
| ........................................................................................GGAGGGAAACCGAAGATGA..................................................... | 19 | 0 | 1 | 4.00 | 4 | 2 | 2 | 0 | 0 | 0 |
| ........................................................................................TGAGGGAAACCGAAGATGA..................................................... | 19 | 1 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 |
| .................................................GTCATCTTCGGTTTTCCTACA.......................................................................................... | 21 | 0 | 1 | 3.00 | 3 | 0 | 0 | 2 | 1 | 0 |
| ........................................................................................GGAGGGAAACCGAAGATGAT.................................................... | 20 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| .....................................................................................TGTGGAGGGAAACCGAAGATGA..................................................... | 22 | 1 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................TGTGGAGGGAAACCTAAGATGA..................................................... | 22 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................GTGGAGGGAAACCGAAGATGA..................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................GGAAACCGAAGATGATGAA................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................GGTTGAGGGAAACCGAAGATGA..................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................TTTGAGGGAAACCGAAGATGA..................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TGGAGGGAAACCGAAGATGA..................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................AGTCATCTTCGGTTTTCCTACA.......................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................GGTGGAGGGAAACCGAAGATGAT.................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................TAACCATTAGTTTTGACGAAGTTT........................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................TGGAGGGAAACGGAAGATGA..................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................TCTTCGGTTTTCCTACACTCAGA.................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TGAGGGAGACCGAAGATGA..................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................AAAAAGTTTAGGTGGAAG................................................................... | 18 | 2 | 20 | 0.25 | 5 | 0 | 0 | 0 | 5 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_12903:550499-550658 + | dan_100 | confident | AAAAAATTTTTAAAAAGTGATAATAATTCTT-----------------------------GGAGTATTATCTAATTAT--CAGTAGAAGCCAAAAGGATGTGAGTATTTTTGAAATCCA----------CCTCCCTTTGGCTTCTACTACTTAATTGGTAATCAAAACTGCTTCAAAAACAGT---------------------CTAAAACTAACTGAAAAG |
| droBip1 | scf7180000395912:19103-19310 + | AAAAAAGTCTTCAAA-----TTAAAGTTCAAAGAGAAGAAAAGTGTTAAAATACAAAAAAAAAGTATTATCTAATTACTACAAGGGGAGTCATAGTGAAATACATATTTTTAAAAACTATTACATTATACCTTCCTTTGG---------CTTAATTGATAATCAAATCTTCAACAAAAAAAATAATAATAATAATAATAATAATAAAAAGTGAAAAATGAAG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 10/20/2015 at 06:45 PM