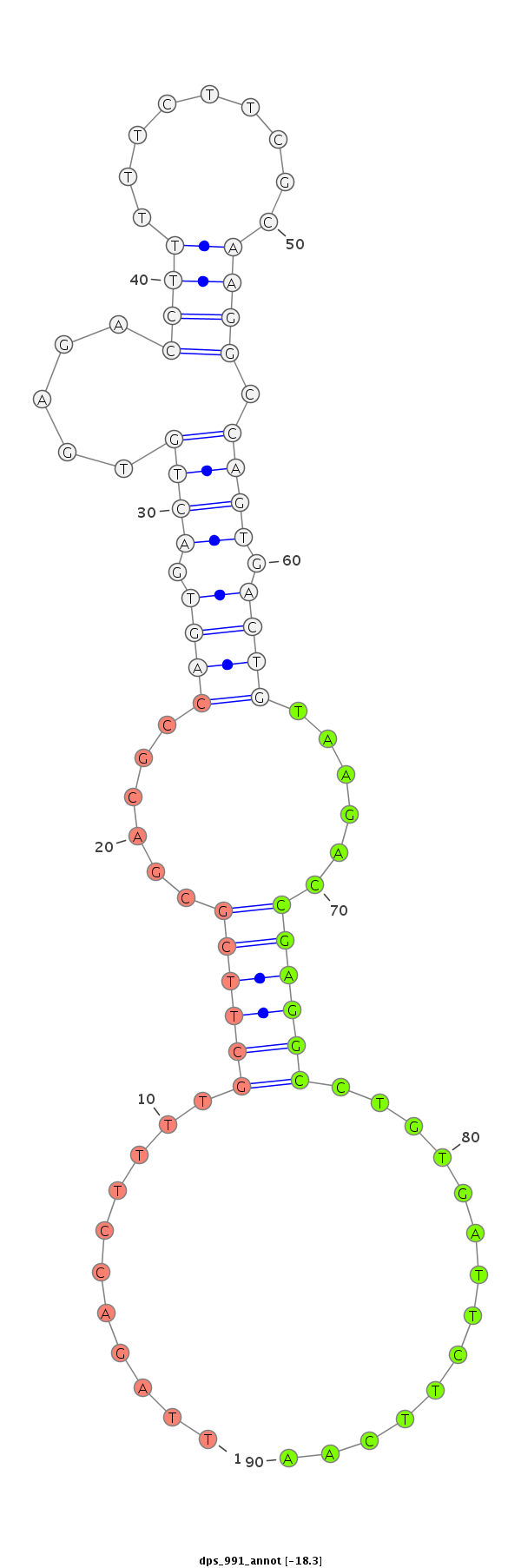
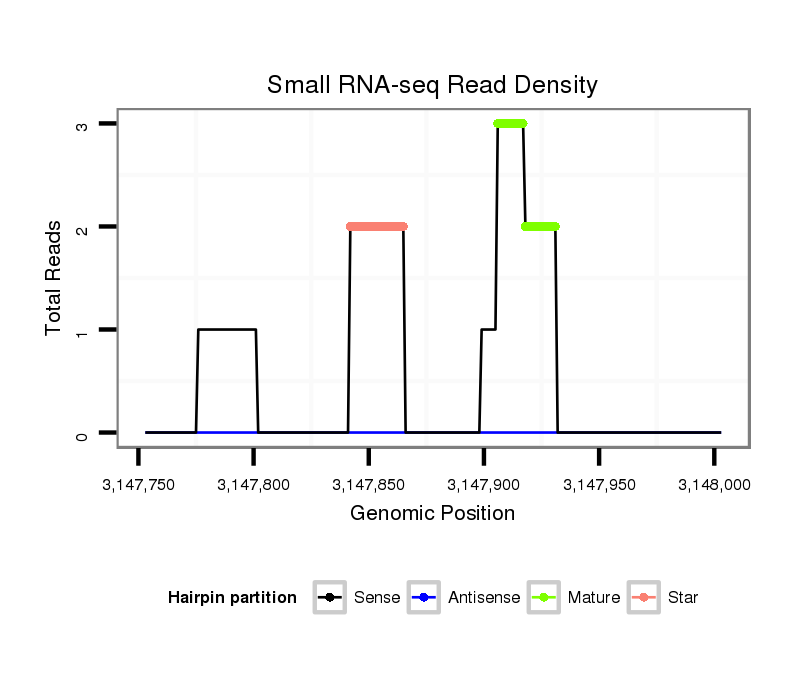
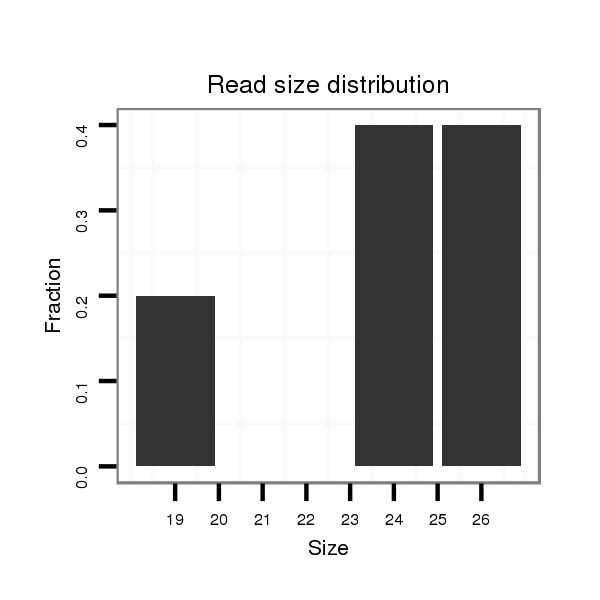
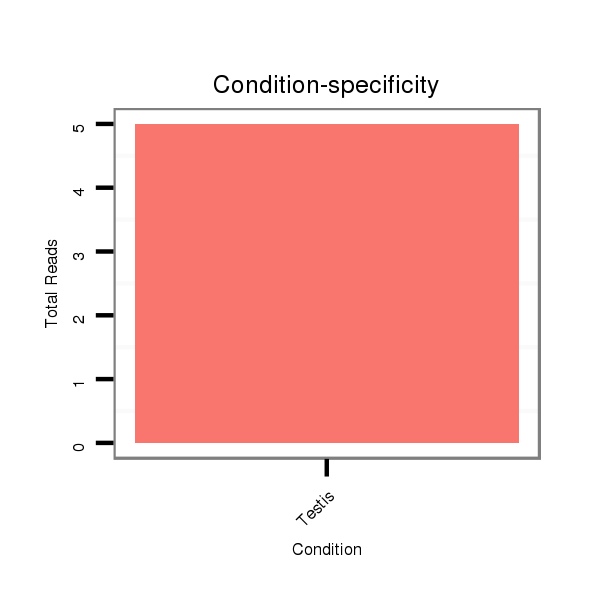
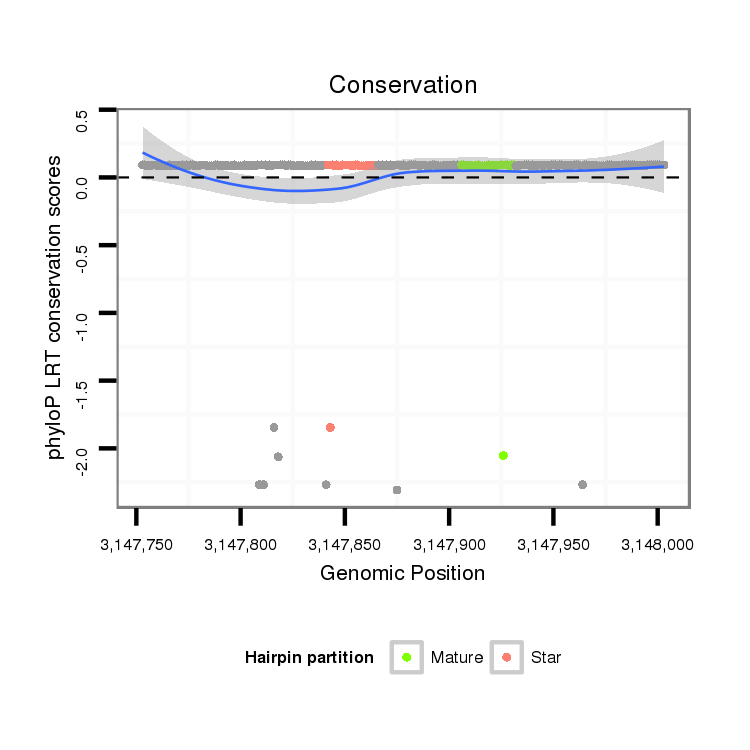
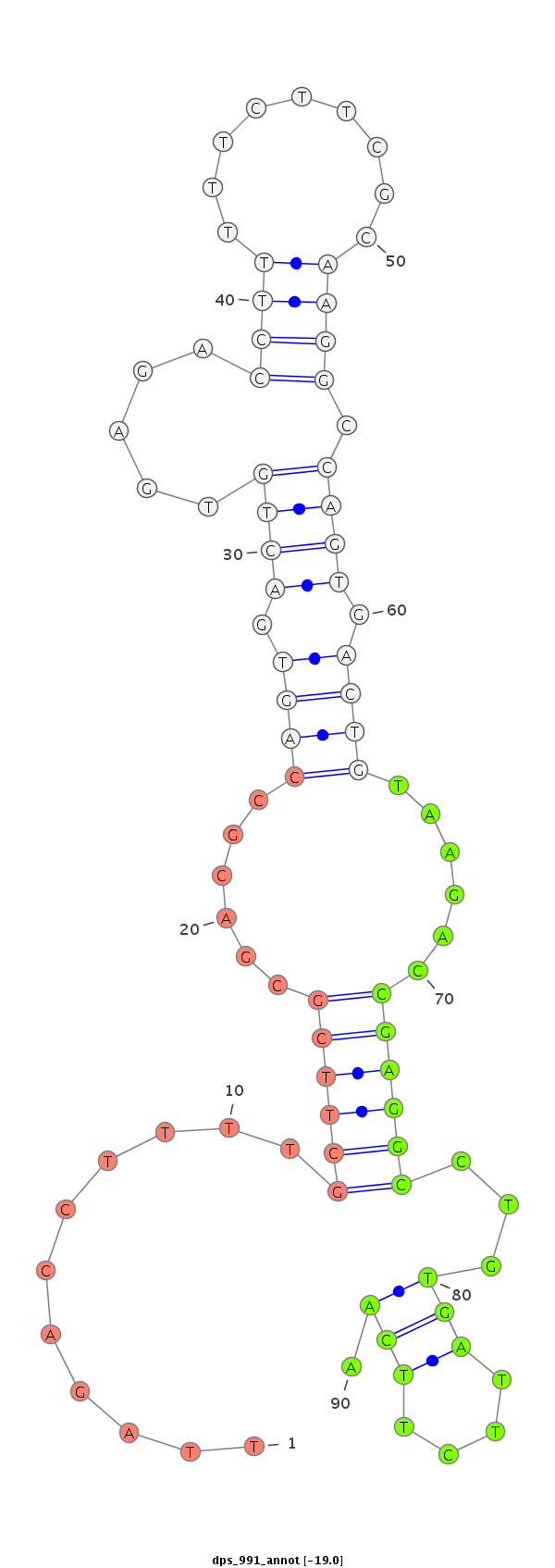
ID:dps_991 |
Coordinate:3:3147803-3147953 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -19.0 | -18.2 | -18.2 |
 |
 |
 |
exon [dpse_GLEANR_15235:2]; CDS [Dpse\GA24668-cds]; intron [Dpse\GA24668-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATCTGTAGCCGGGAGAGCGACAGTACGGATAGCCCTTCCTCGCGAGGCCTGTGACTCTCAGACTTTTTTCATCGCGACGCCAGTGACTCTTAGACCTTTTGCTTCGCGACGCCAGTGACTGTGAGACCTTTTTCTTCGCAAGGCCAGTGACTGTAAGACCGAGGCCTGTGATTCTTCAACCTTTTACTGGATCTACTTTCAATGGCAAATGCTTTCAAGGCCCAAAACTGATTAAAACCAGGATTTAAATT *****************************************************************************************...........((((((......((((.((((.....((((.........)))).)))).))))......))))))..............************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
GSM444067 head |
M040 female body |
|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................TAAGACCGAGGCCTGTGATTCTTCAA........................................................................ | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| .........................................................................................TTAGACCTTTTGCTTCGCGACGCC.......................................................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ..................................................................................................................................................GTGACTGTAAGACCGAGGC...................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................TACGGATAGCCCTTCCTCGCGAGGCC.......................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..............................................................................................................................................................................TTCAACCTTGTTCTGGAT........................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 |
| ...........................................................................................................................................................TGACGGAGGCCTGTGGTTCT............................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 |
|
TAGACATCGGCCCTCTCGCTGTCATGCCTATCGGGAAGGAGCGCTCCGGACACTGAGAGTCTGAAAAAAGTAGCGCTGCGGTCACTGAGAATCTGGAAAACGAAGCGCTGCGGTCACTGACACTCTGGAAAAAGAAGCGTTCCGGTCACTGACATTCTGGCTCCGGACACTAAGAAGTTGGAAAATGACCTAGATGAAAGTTACCGTTTACGAAAGTTCCGGGTTTTGACTAATTTTGGTCCTAAATTTAA
************************************************************************...........((((((......((((.((((.....((((.........)))).)))).))))......))))))..............***************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
SRR902011 testis |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................AGAAGCGTTTCGGTGACTG.................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................CAGGTACCGTTTACGAAAG................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ...............................CGGGAAGGTGCGCTCTGG.......................................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...............................................................AAAAAATTTGCGCTGCGGT......................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ...........................CTACCGGGAAGGTGCGCT.............................................................................................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ............................TACCGGGAAGGTGCGCTCT............................................................................................................................................................................................................ | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................ACAGGAACCGTTTACGAAAG................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ...........................CTAGCGGGAAGGTGCGCC.............................................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ...............................................................AAAACATTTGCGCTGCGGT......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .........................................................................................GGTCTGGAAAACGAAGCGG............................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..............................CCGGGAAGGTGCGCTCTG........................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 3:3147753-3148003 + | dps_991 | ATCTGTAGCCGGGAGAGCGACAGTACGGATAGCCCTTCCTCGCGAGGCCTGTGACTCTCAGACTTTTTTCATCGCGACGCCAGTGACTCTTAGACCTTTTGCTTCGCGACGCCAGTGACTGTGAGACCTTTTTCTTCGCAAGGCCAGTGACTGTAAGACCGAGGCCTGTGATTCTTCAACCTTTTACTGGATCTACTTTCAATGGCAAATGCTTTCAAGGCCCAAAACTGATTAAAACCAGGATTTAAATT |
| droPer2 | scaffold_2:3341281-3341531 + | ATCTGTAGCCGGGAGAGCGACAGTACGGATAGCCCTTCCTCGCGAGGCCTGTGACTGTGAGACCTGTTTCATCGCGACGCCAGTGACTGTCAGACCTTTTGCTTCGCGACGCCAGTGACTGTCAGACCTTTTTCTTCGCAAGGCCAGTGACTGTAAGACCGAGGCCTGTGATTATTCAACCTTTTACTGGATCTACTTTCAATGGCAAATGGTTTCAAGGCCCAAAACTGATTAAAACCAGGATTTAAATT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/17/2015 at 11:46 PM