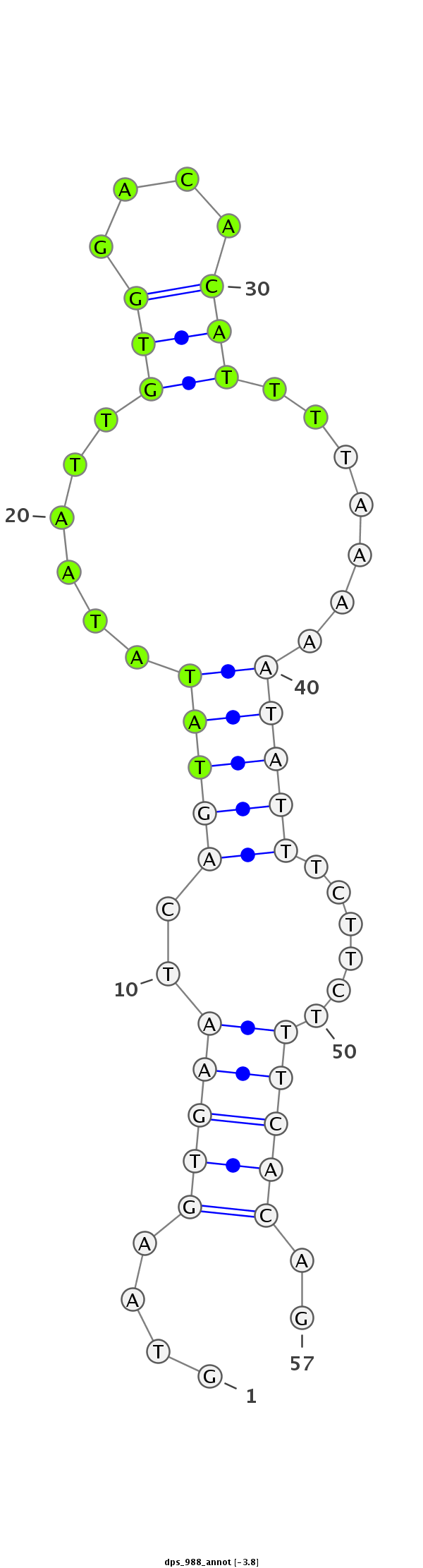
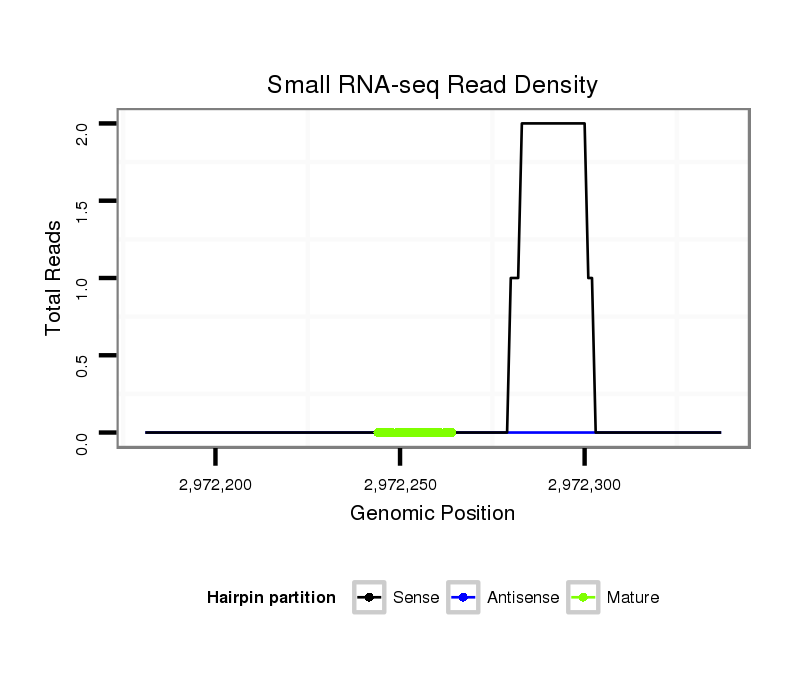
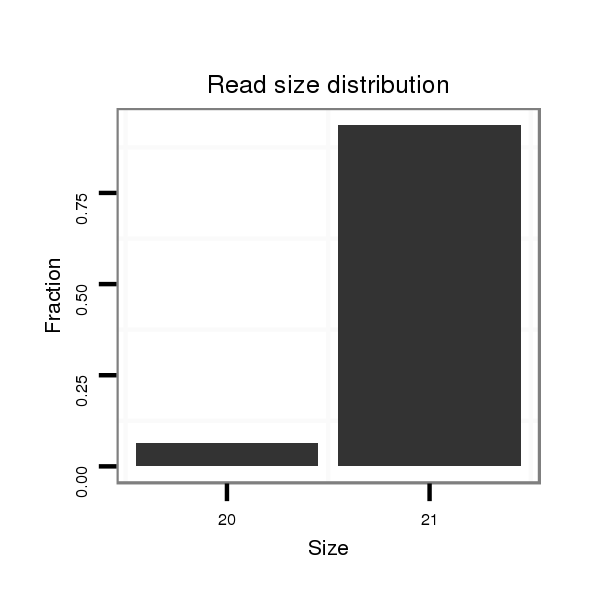
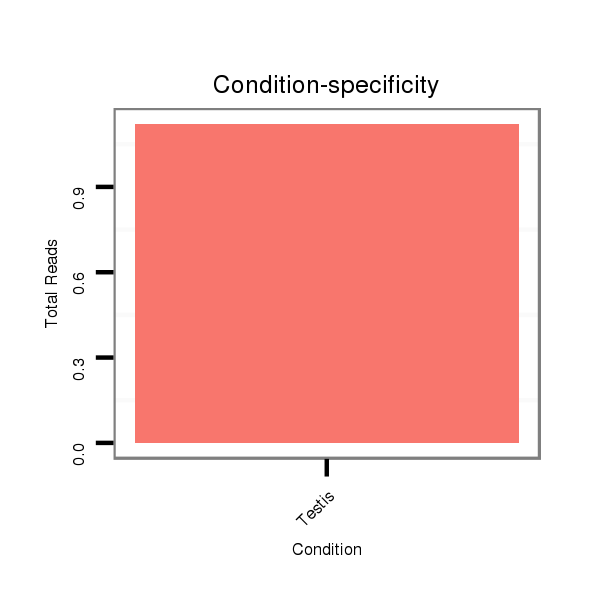
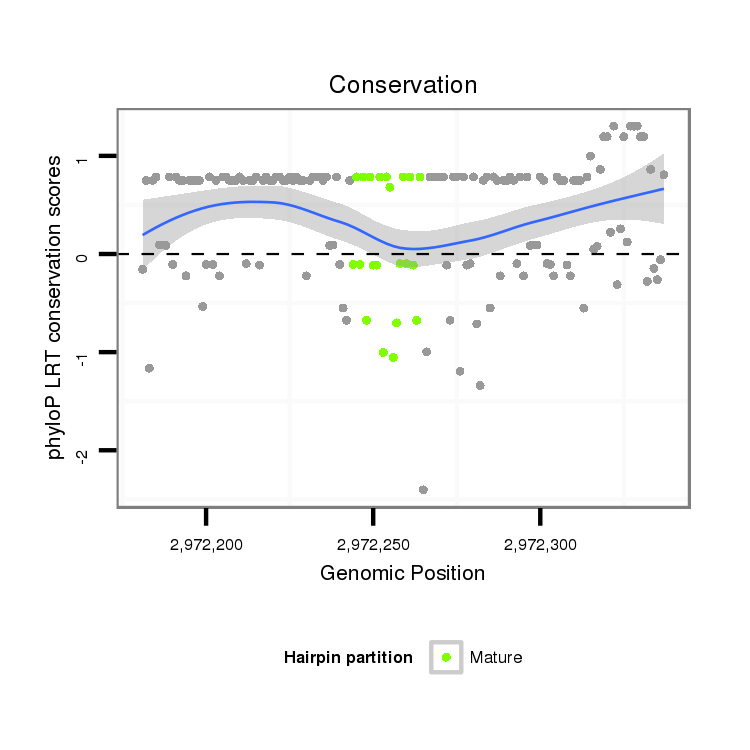
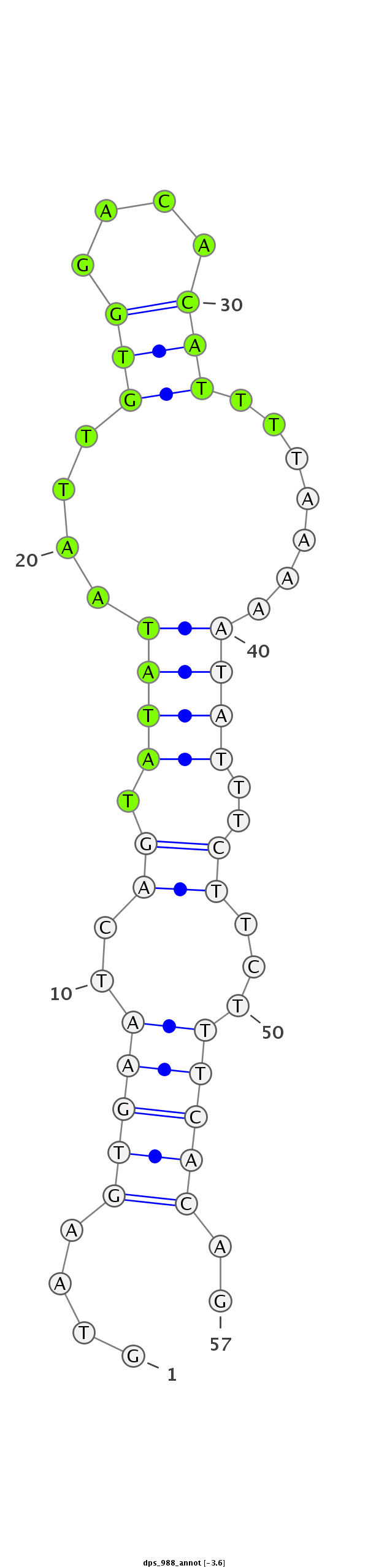
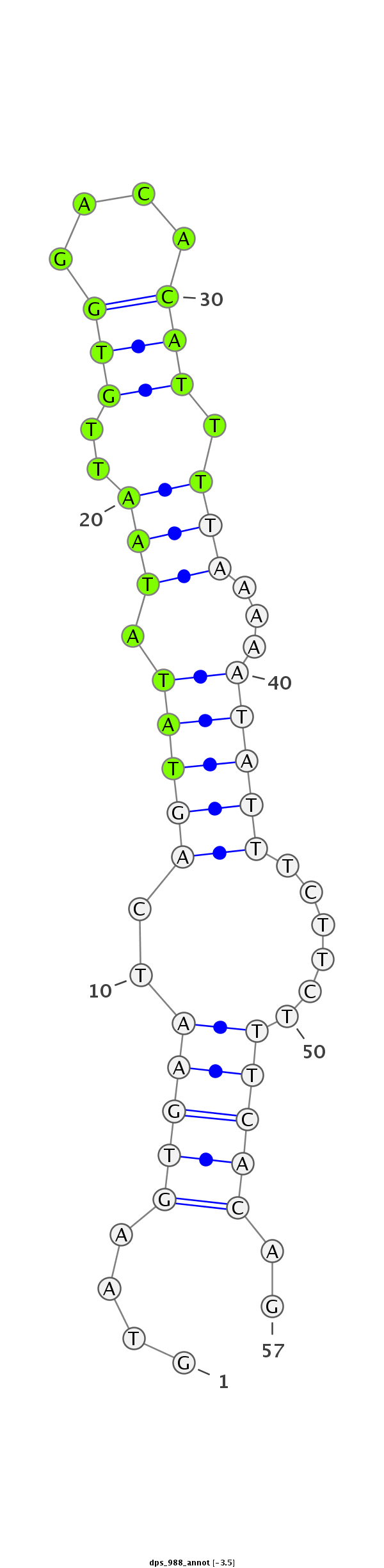
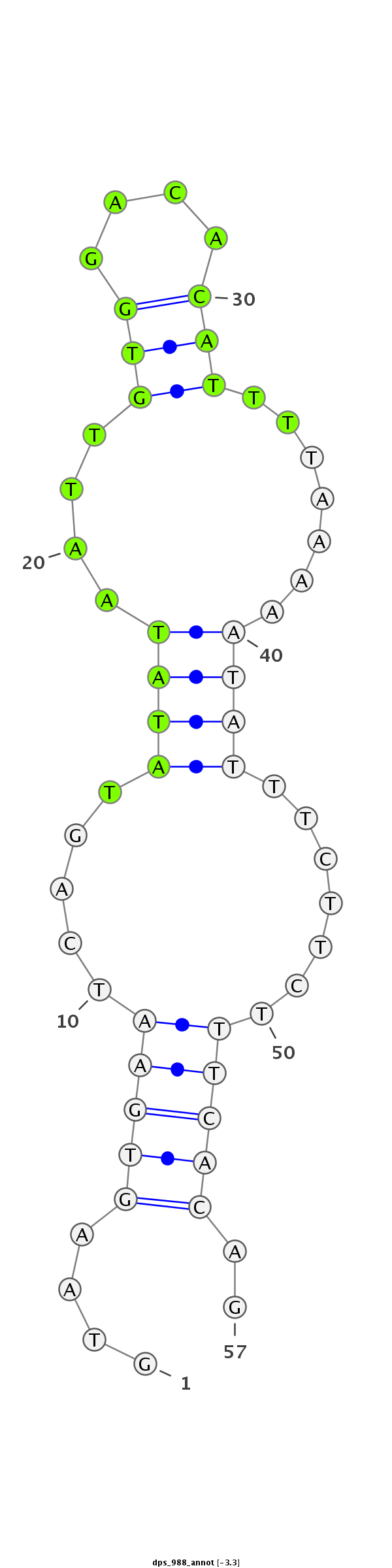
ID:dps_988 |
Coordinate:3:2972231-2972287 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -3.6 | -3.5 | -3.3 |
 |
 |
 |
CDS [Dpse\GA24663-cds]; CDS [Dpse\GA24663-cds]; exon [dpse_GLEANR_15220:2]; exon [dpse_GLEANR_15220:1]; intron [Dpse\GA24663-in]
No Repeatable elements found
| mature | star |
| ##################################################---------------------------------------------------------################################################## GCTCATTCATACGACGCCGAATCAAACGCTGGGCTACAGCTTCGAGACGAGTAAGTGAATCAGTATATAATTGTGGACACATTTTAAAAATATTTCTTCTTTCACAGAGGACCTATGTATCGTAAGCTTCGCCACAAACGAAGTCGTATCCGATTTC **************************************************....(((((..(((((......(((....))).......)))))......)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
V112 male body |
|---|---|---|---|---|---|---|---|
| ......................................................................................................CACAGAGGACCTATGTATCG................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| ...................................................................................................TTTCACAGAGGACCTATGTAT..................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ...............................................................TATATAATTGTGGACACATTT......................................................................... | 21 | 0 | 3 | 0.67 | 2 | 2 | 0 |
| ..................GAATCAAACGCTGGGCTACAG...................................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ........ATACGACGCCGAATCAAACGC................................................................................................................................ | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 |
| .............................TGGGCTACAGCTTCGAGACGAG.......................................................................................................... | 22 | 0 | 3 | 0.33 | 1 | 1 | 0 |
| ..................................TACAGCTTCGAGACGAGTAAG...................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 |
| .............................................................AGTATATAATTGTGGACACAT........................................................................... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 |
| .............................TGGGCTACAGCTTCGAGACGA........................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 |
| ..........................................................ATCAGTATATAATTGTGGAC............................................................................... | 20 | 0 | 14 | 0.07 | 1 | 1 | 0 |
| ..............CGCCGAATCAAACGCTGGG............................................................................................................................ | 19 | 0 | 14 | 0.07 | 1 | 1 | 0 |
| .......................................................TGAATCAGTATATAATTGTGG................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 |
|
CGAGTAAGTATGCTGCGGCTTAGTTTGCGACCCGATGTCGAAGCTCTGCTCATTCACTTAGTCATATATTAACACCTGTGTAAAATTTTTATAAAGAAGAAAGTGTCTCCTGGATACATAGCATTCGAAGCGGTGTTTGCTTCAGCATAGGCTAAAG
**************************************************....(((((..(((((......(((....))).......)))))......)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
GSM444067 head |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................GTGTAAAATTTTTATAAAGAA........................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .GAGTAAGTATGCTGCGGCTTA....................................................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................................................GCATTCGAAGCGGTGTTTGCT................ | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..................................ATGTCGAAGCTCTGCTCATTC...................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ................................CGATGTCGAAGCTCTGCTC.......................................................................................................... | 19 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ...............................CCGATGTCGAAGCTCTGCT........................................................................................................... | 19 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ....................................GTCGAAGCTCTGCTCATTCA..................................................................................................... | 20 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ........................TTGCGACCCGATGTCGGAACTC............................................................................................................... | 22 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| .GGGTAAGTATGGTGCGGCTTA....................................................................................................................................... | 21 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 3:2972181-2972337 + | dps_988 | GCTCATTCATACGACGCCGAATCAAACGCTGGGCTACAGCTTCGAGACGAGTAAGTGAATCAGTATATAATTGTG-------GACACATTTTAAAAATAT-T-TCTTCTTTCACAGAGGACCTATGTATCGTAAGCTTCGCCACAAACGAAGTCGTATCCGAT------TTC |
| droPer2 | scaffold_2:3162649-3162805 + | GCCCATTCATACGACGCCGAATCAAACGCTGGGCTACAGCTTCGAGACGAGTAAGTGAATCAGTATATAATTGTG-------GACACATTTAAAAAATAT-T-TTTTCTTTCACAGAGGACCTATGTATCGTAAGCTTCGCCACAAACGAAGTCGTATCCGAT------TTC | |
| droKik1 | scf7180000302472:1879652-1879665 + | A--------------------------------------------------------------------------------------------------------------------------------------------------CGGAGTCCTATCC------------ | |
| droFic1 | scf7180000454076:109529-109557 - | -----------------------------------------------------------------------------------------------------------------------------------------------CAAACGGAAGCGTATCCCAGCATCACTCC | |
| droEle1 | scf7180000491186:2533027-2533040 + | A--------------------------------------------------------------------------------------------------------------------------------------------------CGGAATCGTATCC------------ | |
| droRho1 | scf7180000777283:2223-2236 - | A--------------------------------------------------------------------------------------------------------------------------------------------------CGGAATCGTATCC------------ | |
| droBia1 | scf7180000302408:3629370-3629392 - | -----------------------------------------------------------------------------------------------------------------------------------------------CAGACGGAATCGTATCCCAG------CCC | |
| droTak1 | scf7180000415880:356792-356805 - | A--------------------------------------------------------------------------------------------------------------------------------------------------CGGAGTCGTATCC------------ | |
| droSim2 | 2l:8938439-8938574 + | GCTCA---ACACGCCGCCGGACCCAACGCTGAGCTTCAGCTTCGAGACGCGTAAGT--ACTGGCACACATATTT--ATTTTGGATATAACTTCAAAATTCCTTTTTATTACCATAGCGGACTTCT---TCACCAGCAGCGCTACGA-------------------------- | |
| droYak3 | 2L:11912940-11913076 + | GCTCA---ACACGCCGCCAGACCCAACGCTGAGCTTCAGCTTCGAGACGCGTAAGT--ACCAGCACATATATATGCATTTTACTTATAATTATAAAATTTGT-TTTATTTGCACAGCGGACTTCT---TCACCAGCAGCGCCACGA-------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/17/2015 at 11:49 PM