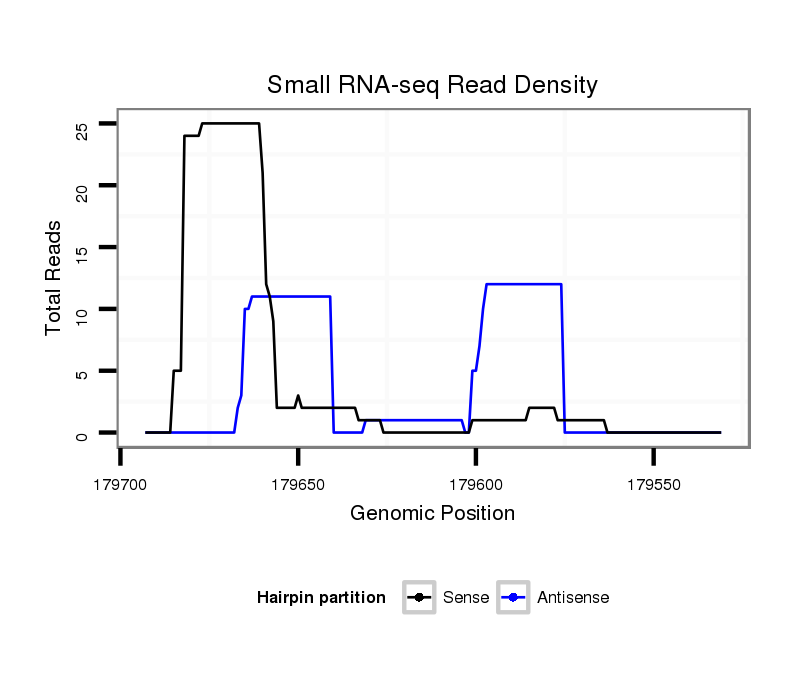
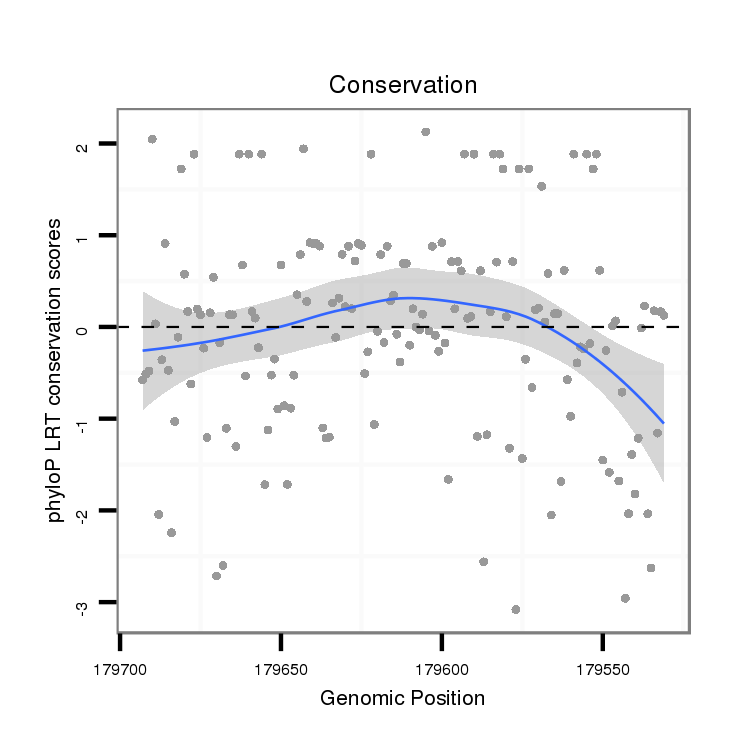
ID:dps_897 |
Coordinate:3:179581-179643 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dpse\GA24586-cds]; exon [dpse_GLEANR_14999:2]; exon [dpse_GLEANR_14999:1]; CDS [Dpse\GA24586-cds]; intron [Dpse\GA24586-in]
| Name | Class | Family | Strand |
| Bilbo | LINE | LOA | - |
| ##################################################---------------------------------------------------------------################################################## AACAATGTTGGTGGATTAGGGACTATGGCATACAAGATCCCAAAATGAAGGTAAAACGAGTACCTGCAAACTTGCAAGACATATCTCTTAAATATCTTTCTGCTCCACAATAGCGATGGCGTAACACGGCGGCAAGCCTAGAGATAAACGGGACTCATCTCAG **************************************************.............((........((..........................))........))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
SRR902010 ovaries |
GSM343916 embryo |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........TGGATTAGGGACTATGGCATACA................................................................................................................................. | 23 | 0 | 1 | 8.00 | 8 | 7 | 1 | 0 | 0 | 0 |
| ...........TGGATTAGGGACTATGGCATACAAGA.............................................................................................................................. | 26 | 0 | 1 | 8.00 | 8 | 6 | 0 | 2 | 0 | 0 |
| ........TGGTGGATTAGGGACTATGGCATAC.................................................................................................................................. | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........TGGATTAGGGACTATGGCATACAAGAA............................................................................................................................. | 27 | 1 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 |
| ...........TGGATTAGGGACTATGGCATAC.................................................................................................................................. | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .....................................TCCCAAAATGAAGGTAAAACGAG....................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................TAGGGACTATGGCATACAAGATCCCAAA....................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........TGGTGGATTAGGGACTATGGCATACAA................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................TATCTTTCTGCTCCACAATAGCGA............................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........TGGTGGATTAGGGACTATGGCATACA................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........TGGATTAGGGACTATGGCATACAAG............................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................AATGAAGGTAAAACGAGTACCTGC................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........TGGTGGATTAGGGACTATGGCATACAAG............................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................AATAGCGATGGCGTAACACGGC................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................TACTTGCAATACATATCGCT........................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................AACACGGCGGCAAGCCTAGAG.................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................TGAAAGATCTTTCTGCGCC......................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TTGTTACAACCACCTAATCCCTGATACCGTATGTTCTAGGGTTTTACTTCCATTTTGCTCATGGACGTTTGAACGTTCTGTATAGAGAATTTATAGAAAGACGAGGTGTTATCGCTACCGCATTGTGCCGCCGTTCGGATCTCTATTTGCCCTGAGTAGAGTC
**************************************************.............((........((..........................))........))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
V112 male body |
SRR902012 CNS imaginal disc |
SRR902010 ovaries |
GSM343916 embryo |
M059 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................GTATGTTCTAGGGTTTTACTTCCAT.............................................................................................................. | 25 | 0 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................ATAGAAAGACGAGGTGTTATCGCTAC............................................. | 26 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GAAAGACGAGGTGTTATCGCTAC............................................. | 23 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................CCGTATGTTCTAGGGTTTTACTTCCAT.............................................................................................................. | 27 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................AGAAAGACGAGGTGTTATCGCTAC............................................. | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................AAAGACGAGGTGTTATCGCTAC............................................. | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CGTATGTTCTAGGGTTTTACTTCCAT.............................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................AAGAATTTATAGAAAGACGAGGT........................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................GGACGTTTGAACGTTCTGTATAGAGAAT......................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................ATGTTCTAGGGTTTTACTTCCAT.............................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................AGATAGGCGAGGTGTTATGG................................................. | 20 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................AAAGAAAGACGAGGGGTTAT................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................AGATAGGCGAGGTGTTATG.................................................. | 19 | 3 | 19 | 0.26 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| ......................................................................................GAAGTTATAGAAAGACGAGGT........................................................ | 21 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TTATAGAAAGACGAGGTGTTA.................................................... | 21 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................CGTATGTTGTAGGGAGTTAC.................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................GTGCCGCCGTTCGGATCTTTATC................ | 23 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................GGGTTTTACTTCCAT.............................................................................................................. | 15 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AGAGTTGATAGAAAGACGACG......................................................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................AGAAAGACGAGGGGTTAT................................................... | 18 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TGTGCCGCCATTCGGATCT..................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TCAGTGCCGCCGTTCGGATCT..................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 3:179531-179693 - | dps_897 | AACAATGTT---GGTGGATTAGGGACTATGGCATACAAGATCCCA--------AAATGAAGGTAAAACGAGTACCTGCAAACTTGCAAGACATATCTCTTAAATATCTTTCTGCTCCACAAT---AGCGATGGCG-TAACACGGCGGCAAGCCTAGAGATAAACGGGA---CT--CATCTCAG |
| droPer2 | scaffold_2:369169-369331 - | AACAAGGTC---GGTGGATTAGGGACTATGGCATACAAGATCCCA--------AAATGAAGGTAAAACGAGTACCTGCAAACTTGCAAGAAATATCTCTTAAATATCTTTCTGCTCCACAAT---AGGGAGGGCG-TAACACGGCGGCAAGCCTAGAGATAAACGGGA---CT--CATCTCAG | |
| droWil2 | scf2_1100000004909:10501717-10501768 - | CTAA-TATG------------------------------------------------ACAGGGAAAGTACATACATACAT----ACAAGACATATCTCTTAAATA------------------------------------------------------------------------------ | |
| droGri2 | scaffold_3023:1569-1741 + | GGCAAGGTT---TGCGGACTAGTAAATAAAGATTATAAGCTCTAAGTAGCTGGAAATGAAGGTAAAAAACGCACCTGCATCCTCGCTAGAAA--ATATTTAAATATTTTTCTGCTTCATAATTTTAGCAATGCCGACAACACAGCGGTAAGCCTGGAGCTCCAGTCAG---CTAC--GGTAAG | |
| droAna3 | scaffold_13417:5387365-5387535 + | GGAAGTGTT---TCTGGACTAGGGACCAGGGAGTTCAAGCTACCAGTAGCTCGCTCTTAAGGTAAAATTAGAGCCTGCATTTTAGCTAAAAA--GCACCTTAGCATATTTTTGCTCCATAATTATAGCGACGCAGAAAA--CGGCAGCTACCGTGGAGTTCCAGACGA---CT-C-TAATTCG | |
| droBip1 | scf7180000391410:12823-12993 - | GACAGGATT---CTTGGGTTAGGGGCGTCGGAGTATAGGCTCTGA--AGCCGATACAACAGGTAAAAGGCGAACCTGCATCCTCACAAAAAA--GCACCTTAGCATCTTTTTGCTACCCAATTTCAGTAATGAAGAGAACTTAGCCGCAACCATAGAGAGCCCAAATG---GCCC--TCTAAG | |
| droKik1 | scf7180000298947:7696-7866 + | AGCAATGCCATCGTCGGAATGAAGTCCGCAAATTACAATTTGCTCCCTCCACCAACGGTAAGCAGGAATAGAACCGCCTTACTTGTACGGAA--GGATCTACACACTAACCTTATGTCCCATTAAAGCTGTGATGACGA--CCGTGGTGATGCTAGAGAGA---GGGA---AA--AAGCGCAT | |
| droEle1 | scf7180000491131:14896-15068 - | GCCAAGATT---TCTGGATTAGGCACTACGGAATACAAGTTATTACTTGGCACAAAAGGTGGTAAAATTCGTACCTGCATTTTAGCCAAAAC--GCACCTTAGCATTTTTCTGCTTCATAATTTTAGCAATGGAGACAGCACCGTGGCAAGTCTGGAACTACCAGCAG---CCAC--GATCAG | |
| droRho1 | scf7180000762034:87-259 - | GGCAGGGTT---GCTGGACTAGGAACAAAAGACTACAAGCTTATGCTTGACCCCAAACAAGGTAAAATTCGAACCTGCATTTTAGCCAAACG--GCATCTTAGTACATTTCTGCTTCGCAATTACAGCAACGGAGACAACACAGCAATAAGCCTGGAGCTGCAAAGCG---GCTC--TTTTAG | |
| droBia1 | scf7180000300606:7955-8121 - | CGA--CATC---TATGCTTTATCCACCCCCTTATACAAGCTATT-------TACACCGAGGGTGAAACCAAAATTTGCATTCTTACCAAGAC--ACACGTTAATGCATATCTCATTCCACAGTTTAGCGAAGGCGACCTCACCACCGTCAGACTGGAGATAAATGAACACACT--CATCACAG | |
| droTak1 | scf7180000412663:6075-6246 - | GGCAAGGTT---GCTGGATTGGGAACAAAAGAGTATAGCCTAT-GCTTGACCCCAAACAAGGTAAAATTCGAACCTGCATTTTAGCCAAAAG--GCTCCTAAATATATTTCTGCTTCGCAATTACAGCAACGGAGATAACACCTCAGTAAGCCTGGAGCTGCAGGACG---GCTC--CATTAG | |
| droYak3 | v2_chr2L_random_010:466888-467060 + | GGAA-GGTT---TCAGGATTAGGGACTAAGGAATATAAGCTTCTAACAGCTCCCAGCGAAGGTAAAATAAGAGCCTGGATTCTAGCCAAAAA--GCACCTTAGCATTTTTCTGCTCCATAATTACAGCGATGCAGACAACGCGGCAGCCACCGTGGAGAGTCAACCAT---CT-CCATTTCGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/17/2015 at 11:32 PM