
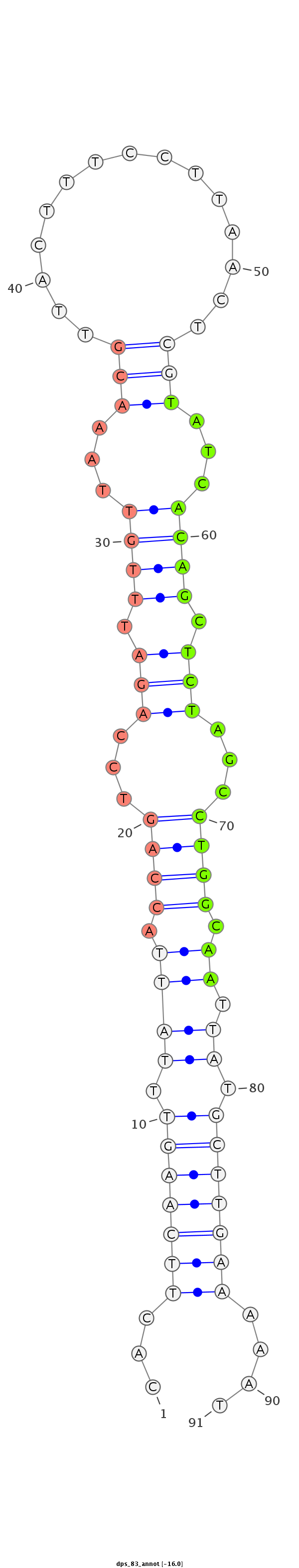
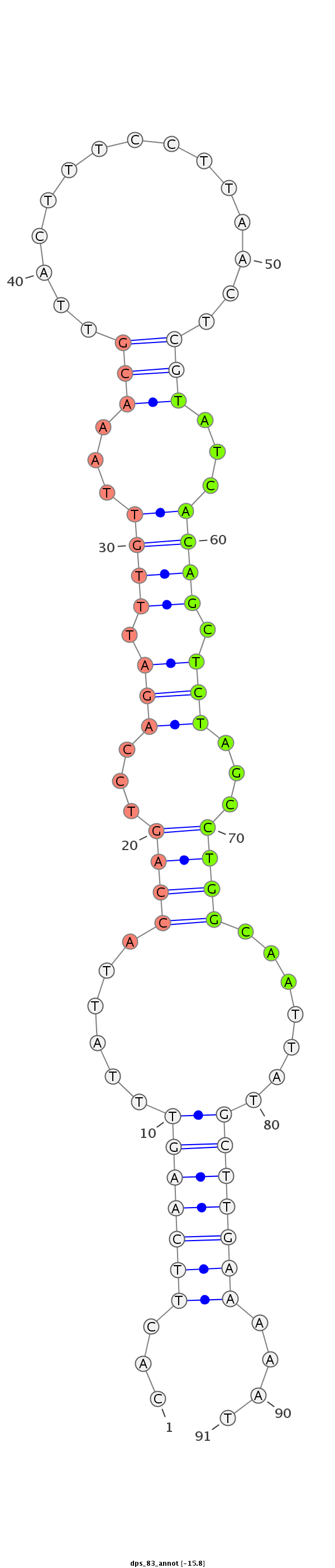
ID:dps_83 |
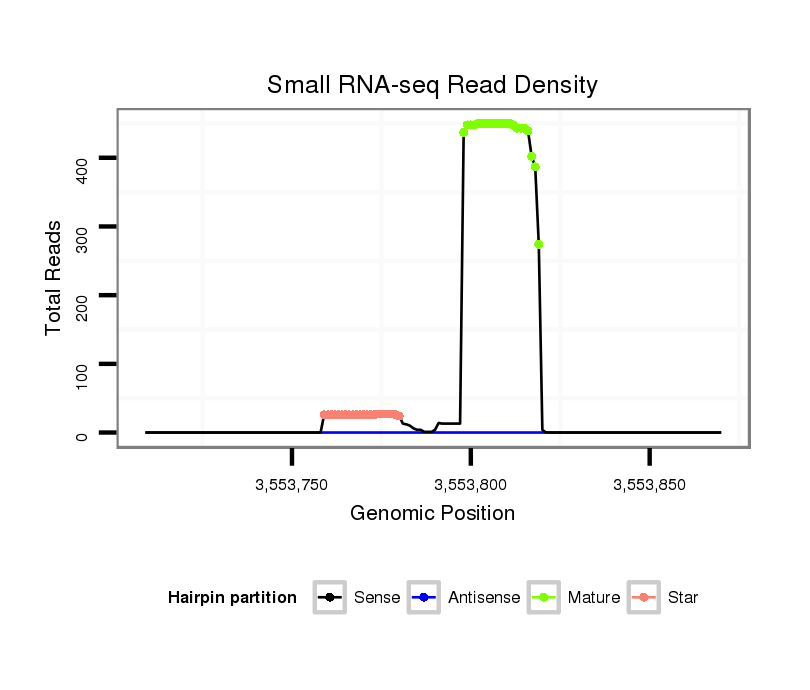
Coordinate:4_group4:3553759-3553820 + |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -16.2 | -16.0 | -15.8 |
 |
 |
 |
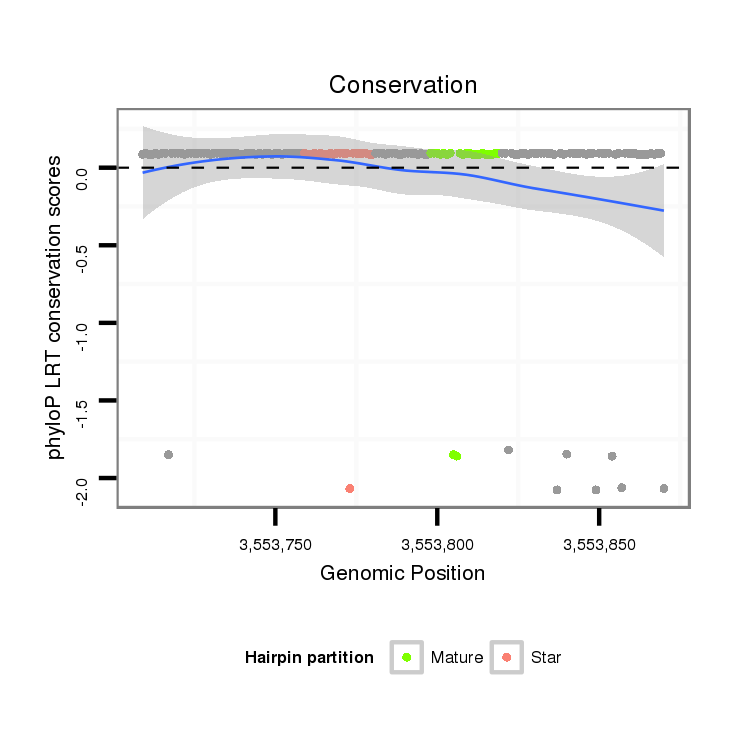
intergenic
No Repeatable elements found
|
CTCCACAAGTAATGATTAAATGAAAATTAAGCAAACACTTCAAGTTTATTACCAGTCCAGATTTGTTAAACGTTACTTTCCTTAACTCGTATCACAGCTCTAGCCTGGCAATTATGCTTGAAAAATACATCTTTTTAGAAAATTTCGATATAAGCGCACAAG
***********************************...(((((((..(((.((((...(((.((((...(((...............)))...)))).)))...)))).)))...)))))))....************************************ |
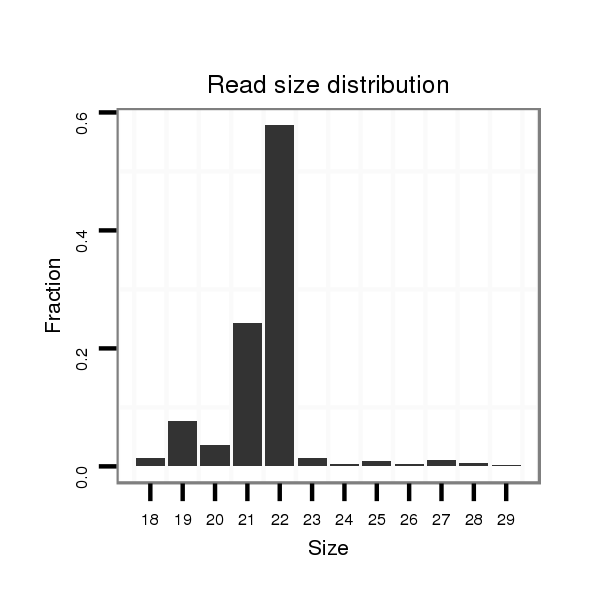
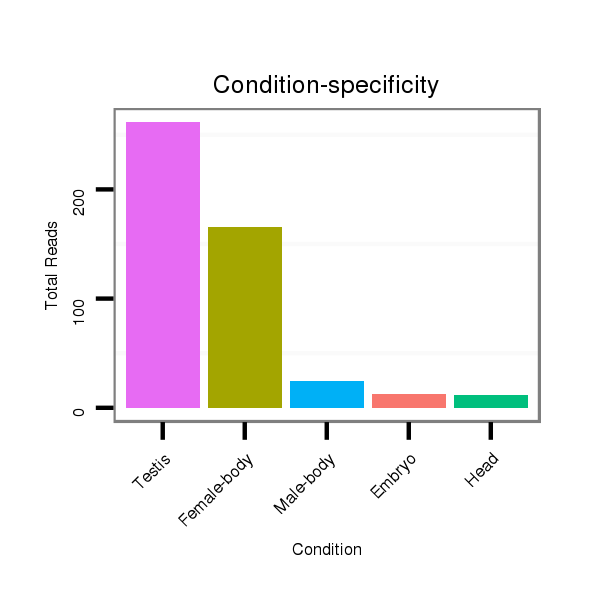
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
M022 male body |
M062 head |
M059 embryo |
V112 male body |
GSM343916 embryo |
GSM444067 head |
SRR902010 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................TATCACAGCTCTAGCCTGGCAA................................................... | 22 | 0 | 1 | 262.00 | 262 | 149 | 91 | 2 | 4 | 9 | 6 | 1 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCA.................................................... | 21 | 0 | 1 | 107.00 | 107 | 61 | 38 | 2 | 4 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGG...................................................... | 19 | 0 | 1 | 37.00 | 37 | 10 | 22 | 4 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAT................................................... | 22 | 1 | 1 | 12.00 | 12 | 10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAAACG.......................................................................................... | 22 | 0 | 1 | 11.00 | 11 | 10 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGC..................................................... | 20 | 0 | 1 | 10.00 | 10 | 3 | 4 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAAA.................................................. | 23 | 1 | 1 | 9.00 | 9 | 7 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................TACCACAGTTCTAACCTGGCAA................................................... | 22 | 3 | 4 | 7.75 | 31 | 6 | 21 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAAAA................................................. | 24 | 2 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATCACAGCTCTAGCCTGGCA.................................................... | 20 | 0 | 1 | 6.00 | 6 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATCACAGCTCTAGCCTGGCAA................................................... | 21 | 0 | 1 | 5.00 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TAACTCGTATCACAGCTCTAGCCTGGC..................................................... | 27 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAAT.................................................. | 23 | 0 | 1 | 4.00 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAAACGTTA....................................................................................... | 25 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTG....................................................... | 18 | 0 | 1 | 4.00 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATTACAGCTCTAGCCTGGCAAAAA................................................ | 25 | 3 | 2 | 3.50 | 7 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................ACCAGTCCAGATTTGTTAAACGTTACTT.................................................................................... | 28 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAAACGTT........................................................................................ | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..................................................................................TAACTCGTATCACAGCTCTAG........................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TAACTCGTATCACAGCTCTAGC.......................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCATA.................................................. | 23 | 2 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAAACGTTAC...................................................................................... | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTAACTCGTATCACAGCTCTAGC.......................................................... | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAAG.................................................. | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGT..................................................... | 20 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTAACTCGTATCACAGCTCTC............................................................ | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAAAC........................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................ACAGCTCTAGCCTGGCAA................................................... | 18 | 0 | 2 | 2.00 | 4 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCATTC................................................. | 24 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAAACGT......................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAACCTGGCAA................................................... | 22 | 1 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATCACAGCTCTAGCCTGGCAAAA................................................. | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAAAAAT............................................... | 26 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCAGGCAA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCC.................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAG............................................................................................. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGA..................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................TAACTCGTACCACAGTTCTAACCTGGCA.................................................... | 28 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TACCACAGCTCTAGCCTGGCAA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAC................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTAACTCGTATCACAGCTCTAG........................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCATTT................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTATAGCCTGGCAA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................ATATCACAGCTCTAGCCTGGCAA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTGGCCTGGCAA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAACA................................................. | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCTTTT................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCAATCC................................................ | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACCAGTCCAGATTTGTTAAA............................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................TTAAACGTTACTTTCCTT............................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCCC................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TAACTCGTATCACAGCTCTAGCCTGGCAA................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCT.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TAGCAAAGCCCTAGCCTGGCAA................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................TTTCACAGCTCTAGCCTGGCAA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAACTCTAGCCTGGCA.................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATTACAGCTCTAGCCTGGCAAGTA................................................ | 25 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TATCACAGCTCTAGCCTGGCATTTT................................................ | 25 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................CAGCTCTAGCCTGGCAAC.................................................. | 18 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGCCTAGAAATTATGCCTGAA........................................ | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GGCAATGATGCTCGAAAACT.................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
GAGGTGTTCATTACTAATTTACTTTTAATTCGTTTGTGAAGTTCAAATAATGGTCAGGTCTAAACAATTTGCAATGAAAGGAATTGAGCATAGTGTCGAGATCGGACCGTTAATACGAACTTTTTATGTAGAAAAATCTTTTAAAGCTATATTCGCGTGTTC
************************************...(((((((..(((.((((...(((.((((...(((...............)))...)))).)))...)))).)))...)))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|
| ................................................AATTGTCCGGCCTAAACAATT............................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 |
| .....................................................TCCGGCCTAAACAATTTGG.......................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 |
| .......................................................................................................................................ATCTTTTACAGCCATATT......... | 18 | 2 | 11 | 0.09 | 1 | 0 | 1 |
| ...................................................................TTCGAAATGATAGGAATTG............................................................................ | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| dp5 | 4_group4:3553709-3553870 + | dps_83 | confident | CTCCACAAGTAATGATTAAATGAAAATTAAGCAAACACTTCAAGTTTATTACCAGTCCAGATTTGTTAAACGTTACTTTCCTTAACTCGTATCACAGCTCTAGCCTGGCAATTATGCTTGAAAAATACATCTTTTTAGAAAATTTCGATATAAGCGCACAAG |
| droPer2 | scaffold_10:2584525-2584686 + | dpe_330 | candidate-rescued | CTCCACAAATAATGATTAAATGAAAATTAAGCAAACACTTCAAGTTTATTACCAGTCCAGATTTTTTAAACGTTACTTTCCTTAACTCGTATCACAATTCTAGCCTGGCAATTTTGCTTGAAAAATACCTCCTTTTAGAACATTTTGAGATAAGCGCACAAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 10/20/2015 at 06:51 PM