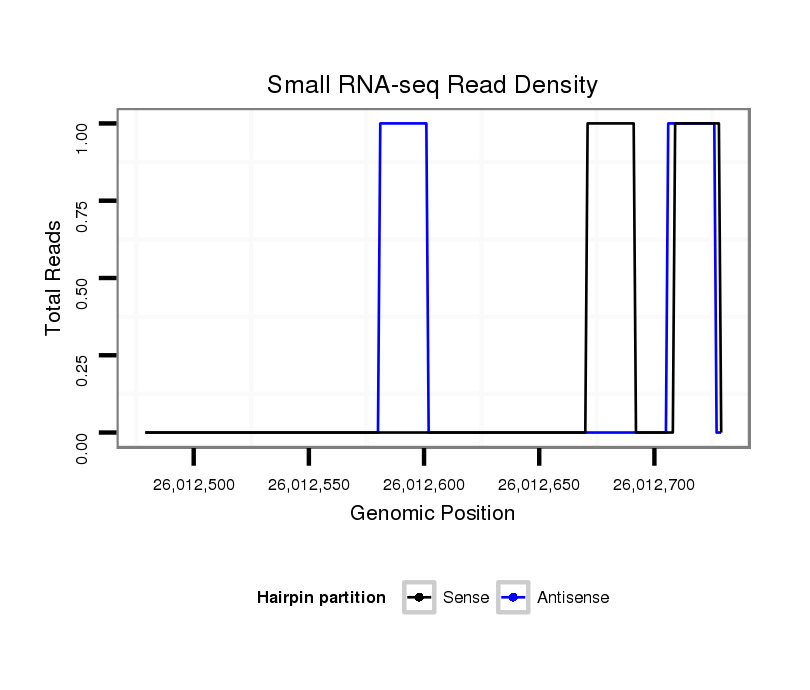
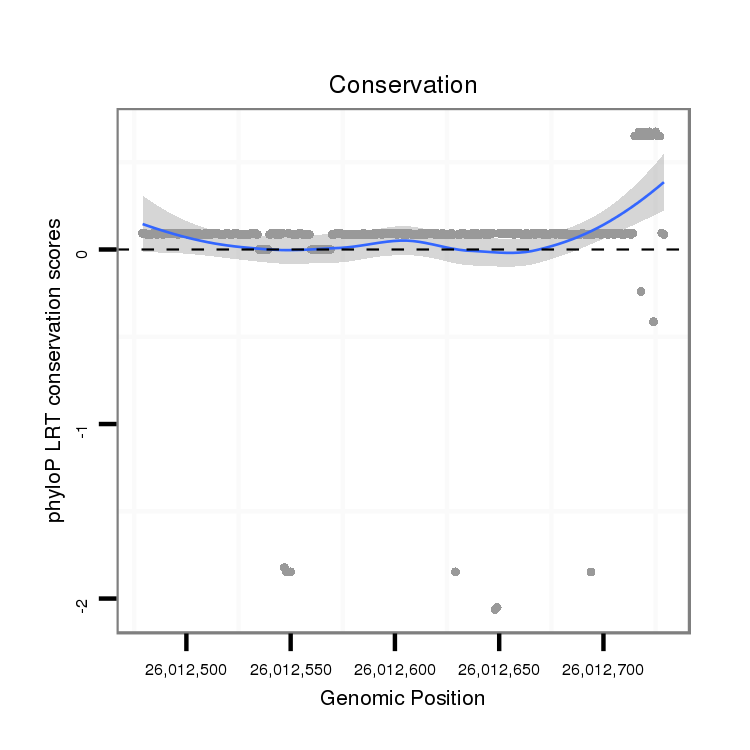
ID:dps_799 |
Coordinate:2:26012529-26012679 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dpse\GA27494-cds]; exon [Dpse\GA27494:5]; intron [Dpse\GA27494-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#################--------------------------------- TTGGCAAGATCATAAAAGAAATCTATGTGCAGAGTTTCCGAAGGTAGGCGGGAATCGAGTTGTTGTGAAAAACCCAAGGGCTTTCCGGGTTGGAATGCCGGCGGCGACGGCAGATCATATTTCAACTCAGCCGACAGTATGTGCACCCTCAGCTTGGCTGAGCTCAGATCGAAATTTTGAAAGCTCCTCTTTCAGCTAGAGTGTGCGCGGGACCCCCAATGTAGATGATAGCTTGAGCATGTGAGATGCAC **************************************************....((((((((..(((...........((((....((((((((((((((.......))))).......)))))))))))))...........)))...))))))))..(((((((((.......)))))..))))((((.......))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
GSM444067 head |
SRR902011 testis |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................CAGCTAGAGTGTGCGCGGGAC...................................... | 21 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GCTTGAGCATGTGAGATGCA. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................GAGTGTGCGTGGGACCCCC.................................. | 19 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TTTCAGCGAGAGTGTTCGCG.......................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................GCACCGTCAGCTTTACTGAGC........................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................TCAGCTAGAGTGTGCGCGGGACCCCCA................................. | 27 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................GTTTGCGCAGGAAGGCGGGAA..................................................................................................................................................................................................... | 21 | 3 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................GCACCTCCAGCTTCGCTGAGCT....................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................CGGGTTCGAATGCCAGCGGCT................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................GCACACTCAGCTTTGATGAGCT....................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................GCACACTCACCTTTGCTGAGCT....................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................TCACCCCCAGCTTTGCTGAGCT....................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................GCACCCCCAGCTTTGATGAGCT....................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ...................................................AATTCGAGTTGTTGTGAA...................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 |
| .........................................................................................................................TGAACACAGCCGACCGTAT............................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................TTCCGGGTTCGAGTCCCGGCG.................................................................................................................................................... | 21 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
|
AACCGTTCTAGTATTTTCTTTAGATACACGTCTCAAAGGCTTCCATCCGCCCTTAGCTCAACAACACTTTTTGGGTTCCCGAAAGGCCCAACCTTACGGCCGCCGCTGCCGTCTAGTATAAAGTTGAGTCGGCTGTCATACACGTGGGAGTCGAACCGACTCGAGTCTAGCTTTAAAACTTTCGAGGAGAAAGTCGATCTCACACGCGCCCTGGGGGTTACATCTACTATCGAACTCGTACACTCTACGTG
**************************************************....((((((((..(((...........((((....((((((((((((((.......))))).......)))))))))))))...........)))...))))))))..(((((((((.......)))))..))))((((.......))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
GSM343916 embryo |
SRR902011 testis |
M022 male body |
M059 embryo |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................TTCGTGGGTTCCGGAAAGGC.................................................................................................................................................................... | 20 | 3 | 13 | 6.77 | 88 | 80 | 6 | 0 | 0 | 1 | 1 | 0 |
| ..........................................................................................................................................TATACGTGGGTGTCGAAGC.............................................................................................. | 19 | 3 | 20 | 2.00 | 40 | 35 | 5 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................CCGCTGCCGTCTAGTATAAAG................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................TATCGAACTCGTACACTCTAC... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TTCTTGGGTTCCGGAAAGGC.................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CCGACTCGAGTCTAGCTTT............................................................................. | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GTTGAGTCGGCTGTCATACAC............................................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................ACTTTCGAGGAGAAAGTCGA...................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ACACGTCTCAAAGGCTTCCAT............................................................................................................................................................................................................. | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................GAGTCGGCTGTCATACACGT.......................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CTGGGGGTTACATCTACTATC.................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................GGCTGTCATACACGTGGGAG..................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....TTCTAGTATTTTCTACAGACACA............................................................................................................................................................................................................................... | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......TCTAGTATTTTCTACAGACACA............................................................................................................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AAACGACTCGAGTGTAGATTT............................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................TAGATAGAGCTCTCAAAGG.................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TCTTGGGTTCCGGAAAGGCG................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................TATACGTGGGTGTCGAAC............................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................AGGATAATGTTGAGCCGG....................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................TCTCGAACCGCCTCGAGTC.................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:26012479-26012729 + | dps_799 | TTGGCAAGATCATAAAAGAAATCTATGTGCAGAGTTTCCGAAGGTAGGCGGGAAT-------------------------------CGAGTTGTTGTGAAAAACCCAAGGGCTTTCCGGGTTGGAATGCCGGCGGCGACGGC---------AGATCATATTTCAACTCAGCCGACAGTATGTGCACCCTCAGCTTGGCTGAGCTCAGATCGAAATTTTGAAAGCTCCTCTTTCAGCTAGAGTGTGCGCGGGACCCCCAATGTAGATGATAGCTTGAGCATGTGAGATGCAC |
| droPer2 | scaffold_6:1344483-1344758 + | TTGGCAAGATCATAAAAGAAATCTATGTGCAGAGTTTCCGAAGGTAGGCGGGAATACTGAGGGGAGGTTGAAGTACGCAAAATATGC-----GTTGTGATGAGCCCAAGGGC----------GGAATGCCGGCGGCGACGGCACAGCTATCAGATCATATTTCAACTCAGCCGACAGTATGTGCACCCTCGGCTTGGCTGAGCTCAGATATAAATTTTGAAAGCTCCTCTTTCAGCTAGAGTGTGCGCGGGACCCTCAATGTAGATGATAGCTTGAGCATGTGAGATGCAC | |
| droTak1 | scf7180000415310:2236-2248 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCACGTGAGCTGC-- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/17/2015 at 11:18 PM