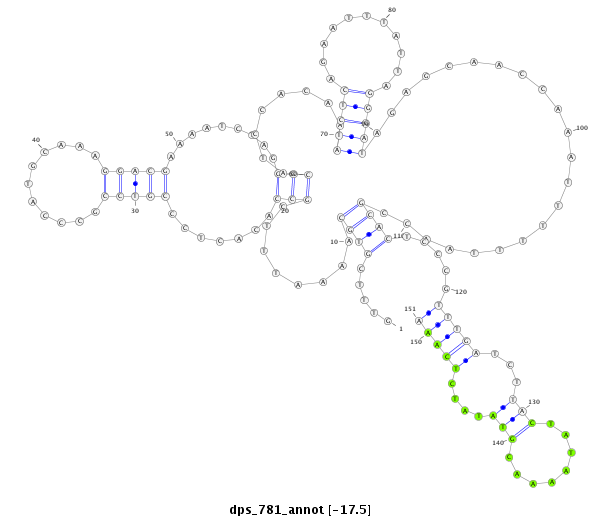



ID:dps_781 |
Coordinate:2:25190221-25190371 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
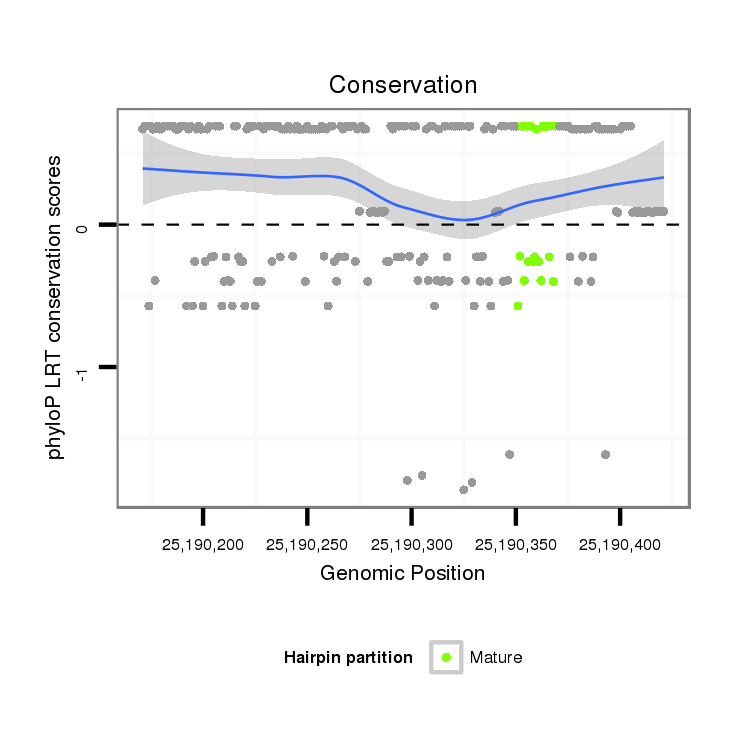
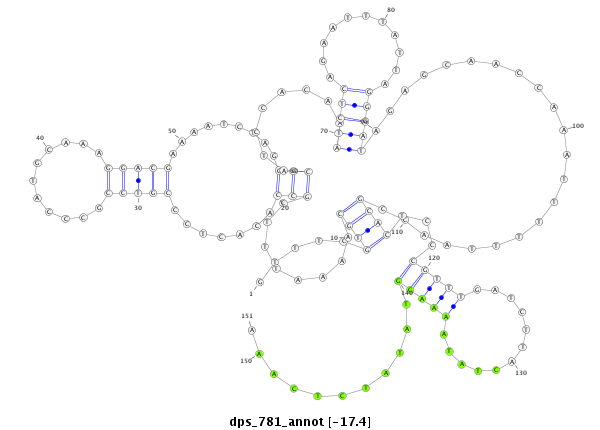
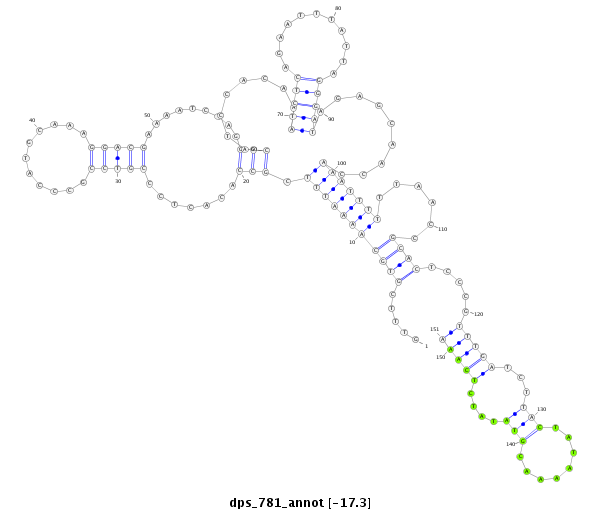
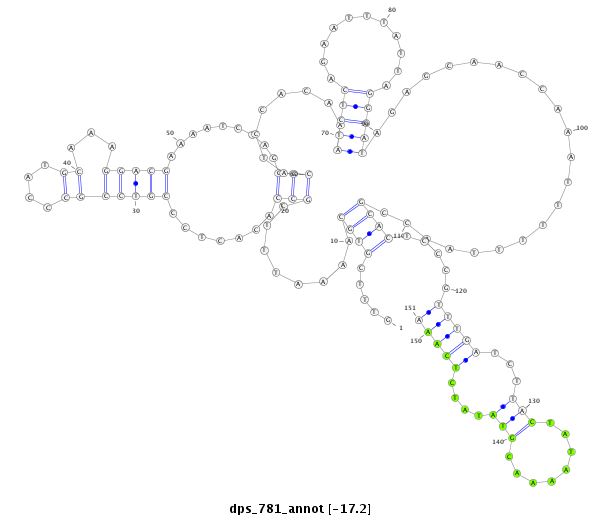
| -17.4 | -17.3 | -17.2 |
 |
 |
 |
CDS [Dpse\GA25584-cds]; exon [Dpse\GA25584:6]; intron [Dpse\GA25584-in]
| Name | Class | Family | Strand |
| Hopers2 | DNA | hAT-hobo | - |
| Hopers2 | DNA | hAT-hobo | - |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTACAGAACCAAATTTGGTATCCACACTCCAGTGATATCGGACCTTGAACGTTTCGTGCAAAATTTCGCCACACTCCCGTCCGCCCATGCAAAGGACGAAAATCTAGGGCATCCACAAATCTCAGAATTTATTAGGGATAGAGCAACCAAATTTTTTAACCGCACTCCCGTTTGATCTTACTATAAAACGTATATCTCAAAATTTCGCCCCACCCCCTTCCGCCCCCACGAAAATCTGTGGCATCCATAAT **************************************************.....((((........(((.......(((((...........))))).........)))........(((((...........)))))......................)))).....(((((...(((........)))....)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
SRR902011 testis |
SRR902010 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................TTATCTTACTATAAAACGTATATC....................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TCTTACTATAAAACGTATATCTCAACATT............................................... | 29 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................TTCGCCCCACCCCCTTCCGCCC.......................... | 22 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 |
| ..........................................................................................AAAGGACGAAAATCTAGGGCATA.......................................................................................................................................... | 23 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................TCTTACCATAAAACGTATATC....................................................... | 21 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................TAAAACGTATATCTCAAAATTTC............................................. | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 |
| ...............................................................................................................................................................................TCTTACTACAAAACGTATA......................................................... | 19 | 1 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ..........AAATTTGGTATCCACACTCCA............................................................................................................................................................................................................................ | 21 | 0 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ........CCAAATTTGGTATCCACACTCCA............................................................................................................................................................................................................................ | 23 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ....................................ATCGGACCATGAACCCTTC.................................................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...............................GTGATATCGGACCCTGAAA......................................................................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TAAAACGTATATCTCAAAATTTAGCCC......................................... | 27 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................CTATAAAACGTATATCTCAA................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GAAAATCTGTGGCATCCAT... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TCGCCCCACCCCCTTCCGCCCCC........................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................CGTATATCTCAAAATTTCGCCC......................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ......AACCAAATTTGGTATCCACACT............................................................................................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....GAACCGAATTTGGTATCCGCACTCC............................................................................................................................................................................................................................. | 25 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................TATCTCAAAATTTCGCCCCACCCC................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................TAAAACGTATATCTCAAAATTTCGCC.......................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................AAACGTATATCTCAAAATTTC............................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................AAAATCTGTGGCATCCAT... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
GATGTCTTGGTTTAAACCATAGGTGTGAGGTCACTATAGCCTGGAACTTGCAAAGCACGTTTTAAAGCGGTGTGAGGGCAGGCGGGTACGTTTCCTGCTTTTAGATCCCGTAGGTGTTTAGAGTCTTAAATAATCCCTATCTCGTTGGTTTAAAAAATTGGCGTGAGGGCAAACTAGAATGATATTTTGCATATAGAGTTTTAAAGCGGGGTGGGGGAAGGCGGGGGTGCTTTTAGACACCGTAGGTATTA
**************************************************.....((((........(((.......(((((...........))))).........)))........(((((...........)))))......................)))).....(((((...(((........)))....)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
V112 male body |
M040 female body |
GSM343916 embryo |
SRR902012 CNS imaginal disc |
M062 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................GGCGTGAGGGCAAACTAGAATGATAT.................................................................. | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................CGTGAGGGCAAACTAGAATGATAT.................................................................. | 24 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................CCGTGAGGGCAAACTAGAATGATAT.................................................................. | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........TTAAACCATAGGTGTGAGGTCACTAT...................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............AAACCATAGGTGTGAGGTCACTAT...................................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................GGTGTGAGGTCACTATAGCCT................................................................................................................................................................................................................. | 21 | 0 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................TGTGAGGTCACTATAGCCTGG............................................................................................................................................................................................................... | 21 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...................TAGGTGTGAGGTCACTATAGC................................................................................................................................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............ACCATAGGTGTGAGGTCACT........................................................................................................................................................................................................................ | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................TAGGTGTGAGGTCACTATAG.................................................................................................................................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GGTGGGGGAAGGCGGGGGTGCTTT.................. | 24 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TGTTTAGGGTCTTAAATATT..................................................................................................................... | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................AATTGGCCTAAGGGAAAACTA........................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................TGGAACTAGCAAAGCATGTA.............................................................................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GGGGAAGGCGGGGGTGCT.................... | 18 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................GTGAGGTCACTCTAGCCTGGAAC............................................................................................................................................................................................................ | 23 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................CCCGTAGGTGTTTAGAGTC.............................................................................................................................. | 19 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 | 0 |
| ..............AACCATAGGTGTTAGTTCCCTAT...................................................................................................................................................................................................................... | 23 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................TCCCGTAGGTGTTTAGAGTCT............................................................................................................................. | 21 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GGGGAGGCGGGGGTGCTT................... | 18 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................CCGTAGGTGTTTAGAGTCTT............................................................................................................................ | 20 | 0 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................TTAGATCCCGTAGGTGTTTAGAG................................................................................................................................ | 23 | 0 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TAGAGTCTTAAACAATAC................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................GGAAGGAGGAGGTGCTTT.................. | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................ATGATATGTTGGATATGGAGT.................................................... | 21 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................GCTTTTAGACACCGTAGGTA... | 20 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TCGTGCTGTTACATCCCGT............................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AGAGTTTTAAAGCGGGGTGGG.................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................GGGGTGGGGGAAGGCGGGGGT....................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CGGGCGGGGGTGCTATTAG............... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................ATATAGAGTTTTAAAGCGGGGT....................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AGAGTTTTAAAGCGGGGTG...................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................GAGTTTTAAAGCGGGGTGG..................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................GTTTTAAAGCGGGGTGGG.................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:25190171-25190421 + | dps_781 | CTACAGA-ACCAAATTTGGTATCCACACTCCAGTGATATCGGACCTTGA-ACGTTTCGTGCAAAATTTCGCCACACTCCCGTCCGCCCATGCAAAGGACGAAAATCTAGGGCATCCACAAATCTCAGAATTTATTAGGGATAGAGCAACCAAATTTTTTAACCGCACTCCCGTTTGATCTTACTATAAAACGTATATCTCAAAATTTCGCCCCACCCCCTTCCGCCCCCACGAAAATCTGTGGCATCCATAAT |
| droPer2 | scaffold_6:496212-496462 + | CTACAGA-ACCAAATTTGGTATCCACACTCCAGTGATATCGGACCTTGA-ACGTTTCGTGCAAAATTTCGCCACACTCCCGTCCGCCCATGCAAAGGACGAAAATCTAGGGCATCCACAAATCTCAGAAATTATTAAGGATAGAGCAACCAAATTTGTTATCCGCACTCCCGTTTGATTTTACTATAAAACGTATATCTCAAAATTTCGCCCCACCCCCTTCCGTCCCCACGAAAATCTGTGGCATCCATAAT | |
| droWil2 | scf2_1100000004849:47679-47901 + | CTAGAGCCACCAAATTTGGTATGCAGTCTCGTGTAGTATGTACAGTTATCTGGTTTGTTTCAAATTTTTGCCACGCTCCCTTCCGCCCACGGAATTAACAAAAAAC-AGGT--------TTTCTTAAAAACTATGTAAGCTACCGACATAAAATTTGGTATGTAAGCTAGC---TTAGCTTAGCAGATATTGACTATTTAAAAATTTTGCCACGCCCATTTCCGCCCCC--GAAAAT---------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/17/2015 at 11:19 PM