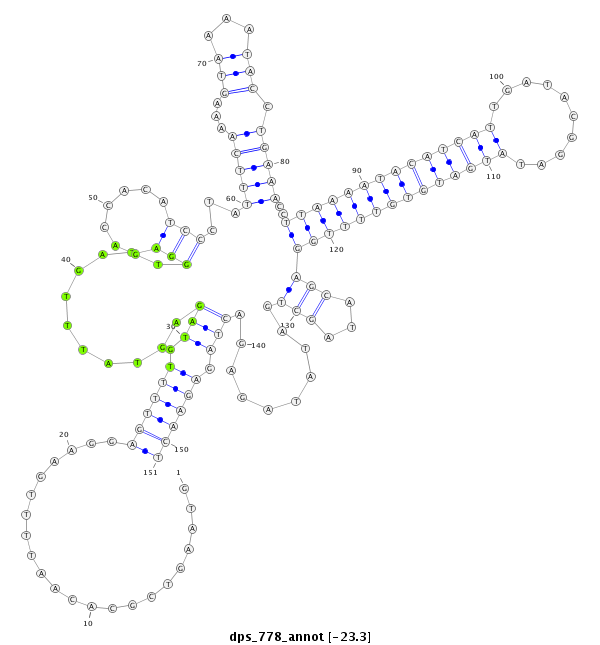
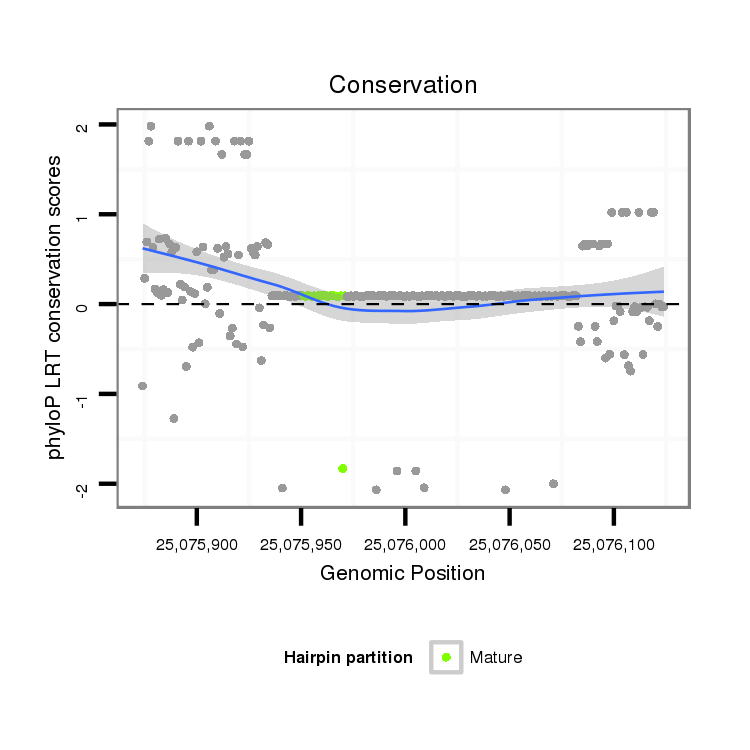
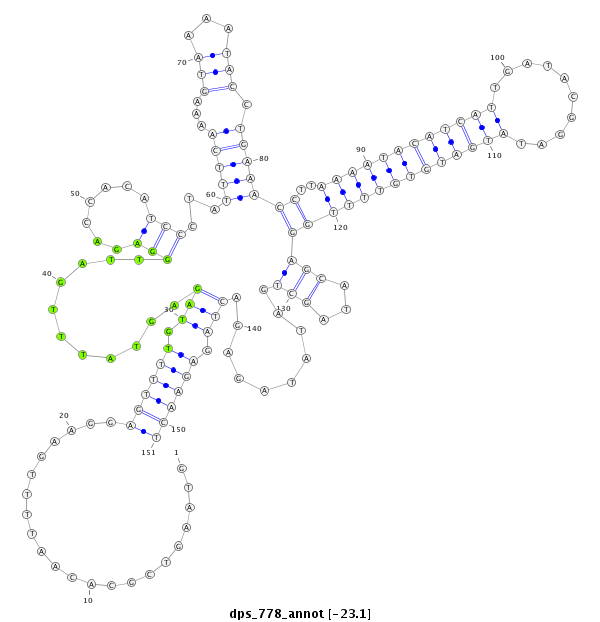
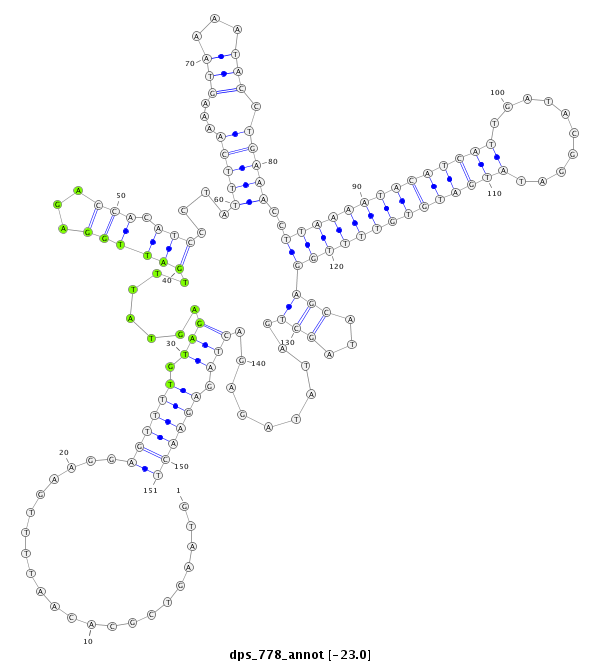
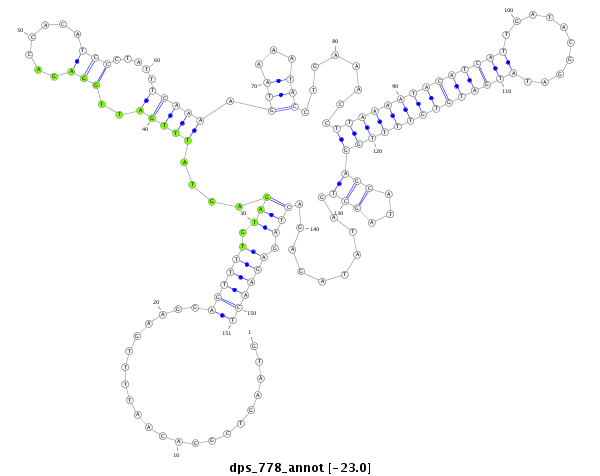
ID:dps_778 |
Coordinate:2:25075924-25076074 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.1 | -23.0 | -23.0 |
 |
 |
 |
CDS [Dpse\GA30211-cds]; exon [Dpse\GA30211:7]; intron [Dpse\GA30211-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CAGCACACTCCTACCACCACTTCTTTCGCGGGACGATAGTTTATTGGAAGGTAAGTCGCACAATTTTGAAGGAGTTTTGTAGAGTATTTGATTGGAGACCACATCCCTATTTCAAAAGTAAAATACCTGAAACCTTAAAATACATCATTGATACGGATATGATGTGTTTTGGAGCATAGCTGATATAGAGACTAGAGAACTTTTCCTTTCACAGAAAGACTTGTATCATCTTTACGACTACTAGATCGGAT **************************************************......................((((((.(((...........(((.......)))...(((((...(((...))).)))))..((((((((((((((..........))))))))))))))(((...)))..........))).))))))************************************************** |
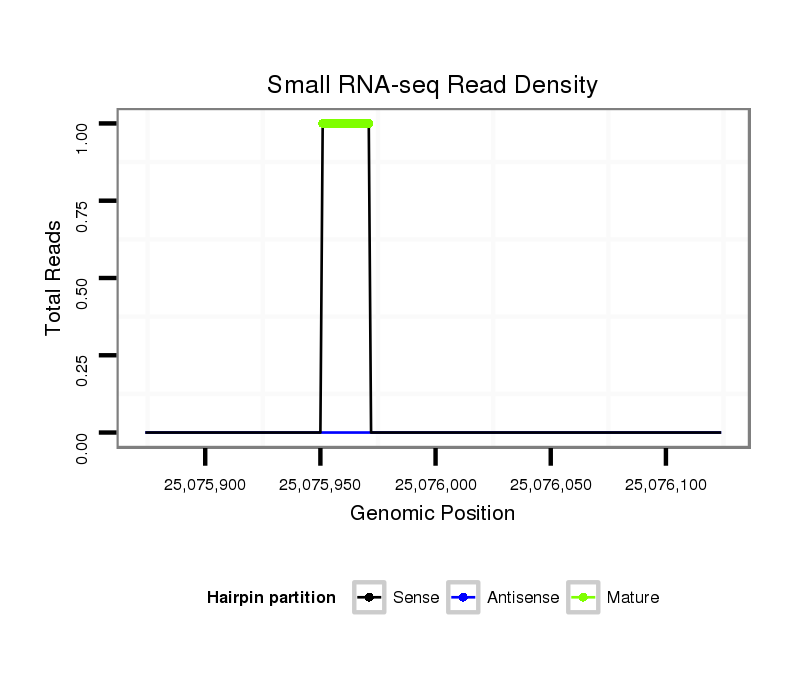
Read size | # Mismatch | Hit Count | Total Norm | Total | V112 male body |
M040 female body |
GSM444067 head |
M059 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................TGTAGAGTATTTGATTGGAGA......................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AGAACTTTTCTTTTCACGGACA.................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .......................TTTGGCCGGAGGATAGTTT................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................GAACTTTTCTTTTCACGGATA.................................. | 21 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................CGCAATTTTGAAGGAGTA............................................................................................................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 |
| .......................................................AAGCGCAATTTTGAAGGAGT................................................................................................................................................................................ | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
GTCGTGTGAGGATGGTGGTGAAGAAAGCGCCCTGCTATCAAATAACCTTCCATTCAGCGTGTTAAAACTTCCTCAAAACATCTCATAAACTAACCTCTGGTGTAGGGATAAAGTTTTCATTTTATGGACTTTGGAATTTTATGTAGTAACTATGCCTATACTACACAAAACCTCGTATCGACTATATCTCTGATCTCTTGAAAAGGAAAGTGTCTTTCTGAACATAGTAGAAATGCTGATGATCTAGCCTA
**************************************************......................((((((.(((...........(((.......)))...(((((...(((...))).)))))..((((((((((((((..........))))))))))))))(((...)))..........))).))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M022 male body |
|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................CAAAGTCGGAATGCTGATGA......... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 |
| ...................................................................................................................................................................................................................GTCTTTCTCAAGATGGTAGA.................... | 20 | 3 | 12 | 0.17 | 2 | 2 | 0 |
| ...GCGGGAGGATGGTGGTGA...................................................................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:25075874-25076124 + | dps_778 | CAGCA-CACTCCT----ACCACC----------ACTTCTTTCGCGGGACGATAGTTTATTGGAAGGTAAGTCGCACAATTTTGAAGGAGTTTTGTAGAGTATTTGATTGGAGACCACATCCCTATTTCAAAAGTAAAATACCTGAAACCTTAAAATACATCATTGATACGGATATGATGTGTTTTGGAGCATAGCTGATATAGAGACTAGAGAACTTTTCCTTTCACAGAAAGACTTGTATCATCTTTACGACTACTAGATCGGAT |
| droPer2 | scaffold_6:375979-376229 + | CAGCA-CACTCCT----ACCACC----------ACTTCTTTCGCGGGACGATAGTTTATTGGAAGGTAAGTCGCACAATTTTTAAGGAGTTTTGTAGAGTATTTGATTGGAAACCACATCCCTATTTAAAAAGTAAAGTACCTGAAGCCTGAAAATACATCATTGATACGGATATGATGTGTTTTGGAGAATAGCTGATATAGAGACTAGAGCACTTTTCCTTTCACAGAAAGACTTGTATCATCTTTACGACTACTAGATCGGAT | |
| droWil2 | scf2_1100000004943:13727132-13727187 + | AAGCA-CACTGCT----TCCGCC----------GCTCCTTTCACGCAACGATGGTTCGCTCGAGGGTAAGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_13047:13518151-13518208 - | GAGCA-CGCTCCT----GCCCCC----------ACTGCTGTCACGCAACGATGGCTCGTTAGAGGGTAAGTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6540:10758236-10758291 - | GAGCA-CACTTCT----GCCCCC----------ACTGCTATCGCGCAACGATGGCTCGTTAGAAGGTAAGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15074:504503-504591 + | GAGCA-CACTCCT----GCCGCC----------TCTACTGTCACGCAACGATGGCACCCTGGAAGGTAAGCTCTACT------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTGCATATCTTTATATAAAATTCGTA | |
| droAna3 | scaffold_12911:1781838-1781897 - | CGTCA-AACTCTTATGTGCCTAC----------AGATCCTACCCGAGATAAAAGTTCGTTAAAAGGTGAAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000391950:13475-13519 + | TTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCAGAAAACCTTCTTTTATCTGTGGGACTGATGAATCGAAT | |
| droRho1 | scf7180000779466:11765-11820 + | AAGCA-CTTACTT----GCCTCC----------ATATCGCGCGCAGAATAATCGTTCACTAGACGGTACGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEug1 | scf7180000409581:29303-29346 - | CAGCATCAGTCCG----GTGACCAATCAGACTAACTCCTTTGGCGGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/17/2015 at 11:16 PM