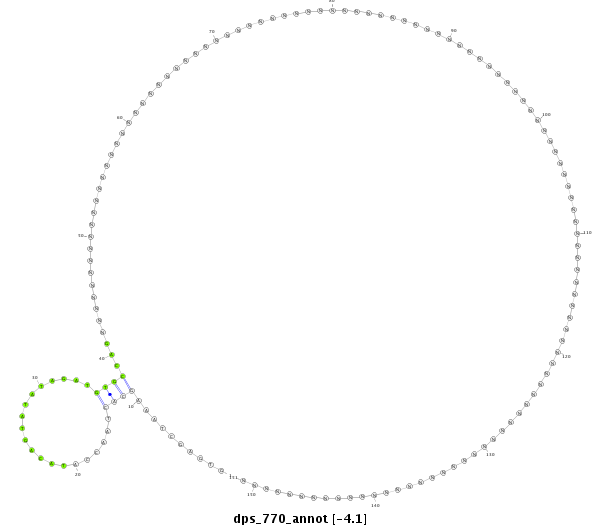
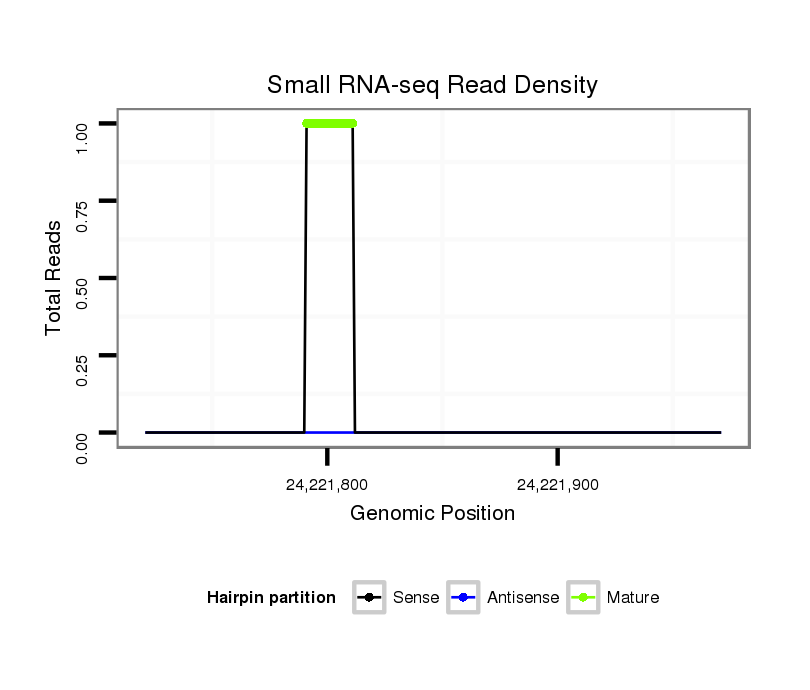
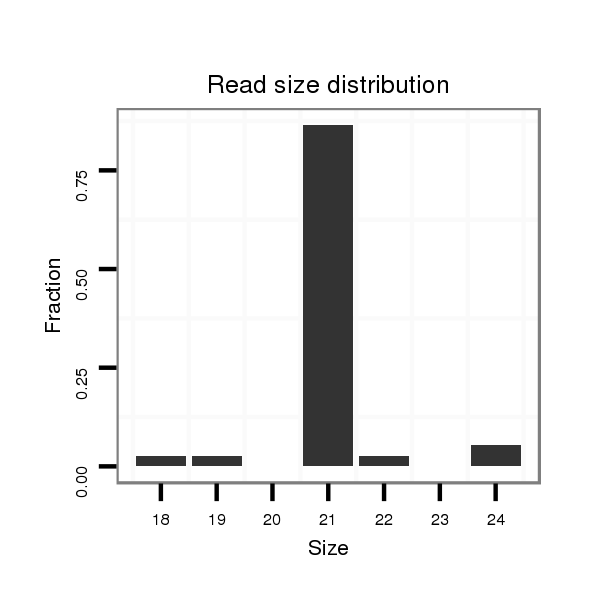
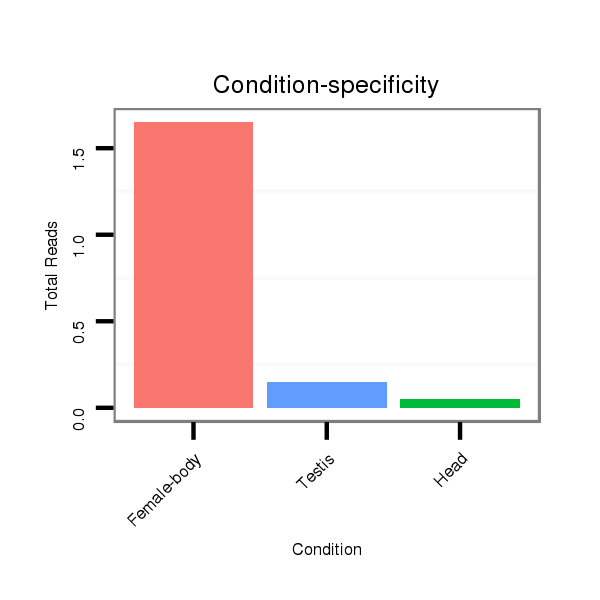
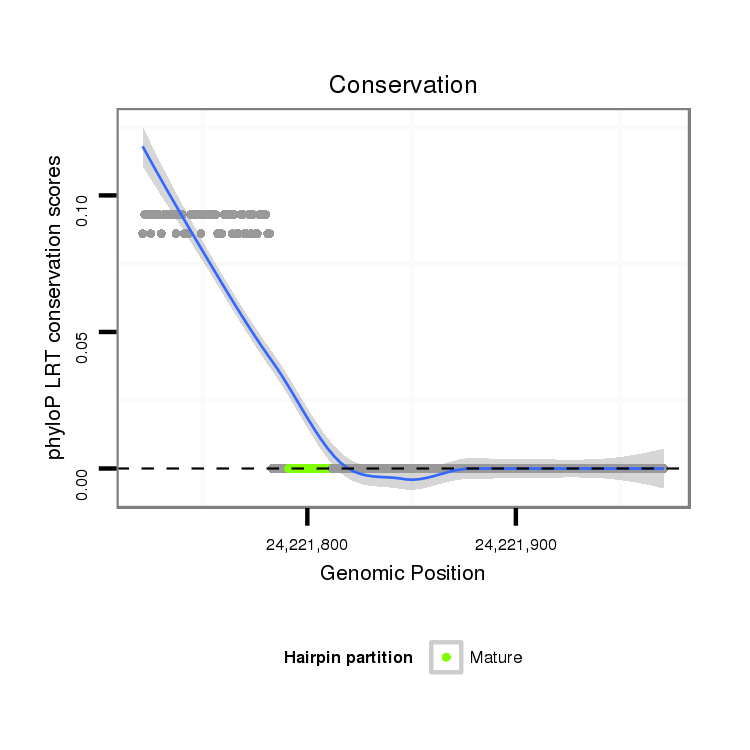
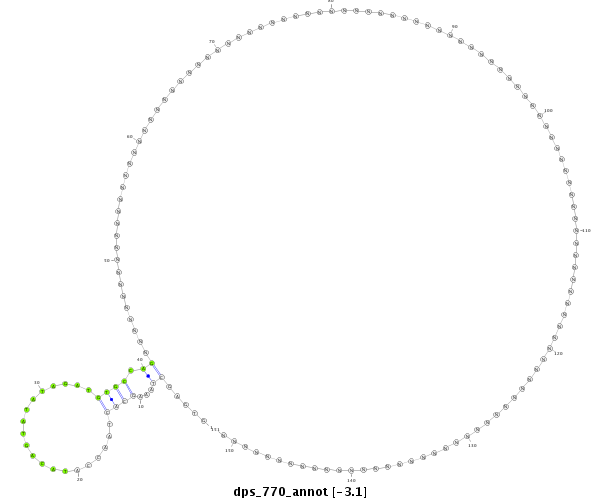
ID:dps_770 |
Coordinate:2:24221771-24221921 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -3.1 |
 |
exon [Dpse\GA27462:1]; CDS [Dpse\GA27462-cds]; intron [Dpse\GA27462-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GATTCAAAAGATATTAGATTGCCAATTTGAAATATTCGGTATTGTGCAAGGTGAGCTAAAGCACTAACCATACAGTATATAGATGTGCCAGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN **************************************************..........((((....................)))).................................................................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
M059 embryo |
M062 head |
|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TACAGTATATAGATGTGCCAG................................................................................................................................................................ | 21 | 0 | 20 | 1.45 | 29 | 26 | 3 | 0 | 0 |
| ................................................................TAACCATACAGTATATAGATGTGC................................................................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................ATACAGTATATAGATGTGCCA................................................................................................................................................................. | 21 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................ATACAGTATATAGATGTGC................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ................................................................TAACCATACAGTATATAGATG...................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................TGCAGTATATAGATGTGCC.................................................................................................................................................................. | 19 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................AGCACTAACCATACAGTATATA.......................................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................AGTATATAGATGTGCCAG................................................................................................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................CATACAGTATATAGATGTGCCC................................................................................................................................................................. | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
CTAAGTTTTCTATAATCTAACGGTTAAACTTTATAAGCCATAACACGTTCCACTCGATTTCGTGATTGGTATGTCATATATCTACACGGTCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
**************************************************..........((((....................)))).................................................................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
|---|---|---|---|---|---|---|---|
| ....................................................................GTATGTCATATATCTACACGG.................................................................................................................................................................. | 21 | 0 | 20 | 0.25 | 5 | 5 | 0 |
| ..........................................................TTCGTGATTGGTATGTCATATAT.......................................................................................................................................................................... | 23 | 0 | 9 | 0.11 | 1 | 0 | 1 |
| ...............................................................GATTGGTATGTCATATATCTACACGGT................................................................................................................................................................. | 27 | 0 | 20 | 0.10 | 2 | 0 | 2 |
| ................................................................ATTGGTATGTCATATATCTAC...................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:24221721-24221971 + | dps_770 | GATTCAAAAGATATTAGATTGCCAATTTGAAATATTCGGTATTGTGCAAGGTGAGCTAAAGCACTAACCATACAGTATATAGATGTGCCAGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN |
| droPer2 | scaffold_3:6991601-6991662 + | GATTCAAAAGATATTAGATTGCCAATTTGAAATATTCGGTATTGTGCAAGGTGAGCTAAAGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/17/2015 at 11:17 PM