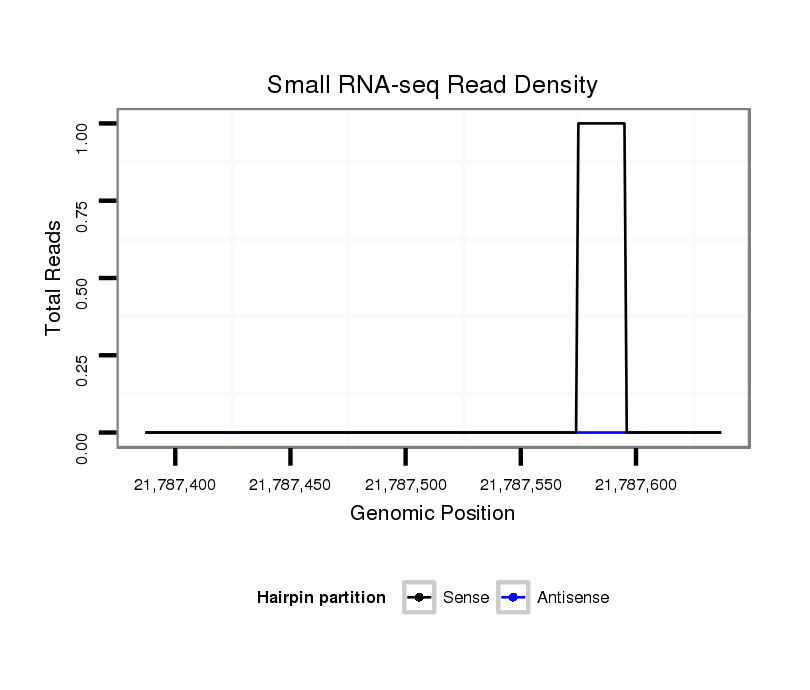
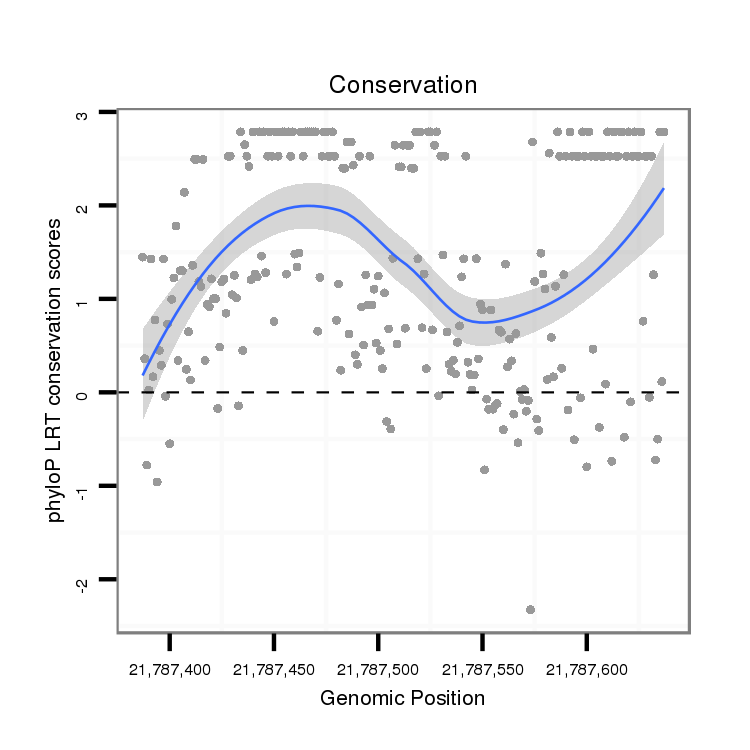
| .............................................................................................................................................ACGTGGTGGTGGGTGTTTT........................................................................................... |
19 |
2 |
14 |
471.57 |
6602 |
5739 |
861 |
0 |
0 |
0 |
1 |
1 |
| .............................................................................................................................................ACGTGTTGGTGGGTGTTT............................................................................................ |
18 |
1 |
1 |
20.00 |
20 |
16 |
4 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTTGTGGGTGTTT............................................................................................ |
18 |
1 |
1 |
16.00 |
16 |
16 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGTTGGTGGGTGTTTT........................................................................................... |
19 |
1 |
1 |
11.00 |
11 |
6 |
5 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGTTGGTGGGTGTTTT........................................................................................... |
18 |
1 |
2 |
10.50 |
21 |
21 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGCTGGTGGGTGTTTT........................................................................................... |
19 |
2 |
2 |
7.00 |
14 |
8 |
6 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGGTTGTGGGTGTTT............................................................................................ |
19 |
2 |
11 |
6.09 |
67 |
33 |
34 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGCTGGTGGGTGTTT............................................................................................ |
18 |
2 |
7 |
5.71 |
40 |
35 |
5 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGTTGGTGGGTGTTTT........................................................................................... |
20 |
2 |
2 |
4.50 |
9 |
6 |
3 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGGTTGTGGGTGTTTT........................................................................................... |
18 |
1 |
5 |
4.20 |
21 |
21 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGGTTGTGGGTGTTTT........................................................................................... |
20 |
2 |
6 |
4.00 |
24 |
10 |
14 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGTTGGTGGGTGTTT............................................................................................ |
19 |
2 |
10 |
3.90 |
39 |
25 |
14 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGCTGGTGGGTGTTTT........................................................................................... |
20 |
3 |
17 |
2.88 |
49 |
32 |
17 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGATGGTGGGTGTTTT........................................................................................... |
19 |
2 |
2 |
2.50 |
5 |
2 |
3 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGATGGTGGGTGTTT............................................................................................ |
19 |
3 |
20 |
2.05 |
41 |
16 |
25 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTTGTGGGTGTTTT........................................................................................... |
19 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGTTGGTGGGTGTTTTAG......................................................................................... |
21 |
2 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGGTGGTGGGTGTTTTT.......................................................................................... |
19 |
2 |
20 |
1.75 |
35 |
34 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGTTGGTGGGTGTTTTA.......................................................................................... |
19 |
2 |
5 |
1.60 |
8 |
6 |
2 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGTTGGTGGGTGTTTTAG......................................................................................... |
20 |
2 |
2 |
1.50 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGATGGTGGGTGTTT............................................................................................ |
18 |
2 |
8 |
1.38 |
11 |
10 |
1 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................AACGTGGTGGTGGGTGTTTT........................................................................................... |
20 |
2 |
4 |
1.25 |
5 |
1 |
4 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTGGTGGGTGTTTTT.......................................................................................... |
20 |
2 |
7 |
0.86 |
6 |
2 |
4 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................AACGTGGTGGTGGGTGTTT............................................................................................ |
19 |
2 |
6 |
0.83 |
5 |
3 |
2 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGATGGTGGGTGTTTT........................................................................................... |
20 |
3 |
20 |
0.75 |
15 |
5 |
10 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGCTGGTGGGTGTTTTAGT........................................................................................ |
21 |
3 |
4 |
0.75 |
3 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTGGTGGGTGTTTTTG......................................................................................... |
21 |
2 |
3 |
0.67 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTTGTGGGTGTTTTA.......................................................................................... |
20 |
2 |
3 |
0.67 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGGTGGTGGGTGTTTTTG......................................................................................... |
22 |
3 |
8 |
0.63 |
5 |
2 |
2 |
0 |
0 |
1 |
0 |
0 |
| ............................................................................................................................................TACGTGGTGGTGGGTGTTTTTGT........................................................................................ |
23 |
3 |
5 |
0.60 |
3 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................ATACGTGGTGGTGGGTGTTT............................................................................................ |
20 |
3 |
20 |
0.45 |
9 |
8 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGCTGGTGGGTGTTTTAG......................................................................................... |
20 |
3 |
16 |
0.38 |
6 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................TTAGCCAGCGGGTGGAGGGGG............................... |
21 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGTTGGTGGGTGTTTTA.......................................................................................... |
20 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTGGTGGGTGTTTTTGT........................................................................................ |
22 |
2 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGTTGGTGGGTGTTTTA.......................................................................................... |
21 |
3 |
13 |
0.31 |
4 |
1 |
3 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTAGTGGGTGTTT............................................................................................ |
18 |
2 |
14 |
0.29 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGGTGGTGGGTGTTTTTG......................................................................................... |
20 |
2 |
7 |
0.29 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................TACGTGTTGGTGGGTGTTTTAG......................................................................................... |
22 |
3 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................ATACGTGGTGGTGGGTGTTTT........................................................................................... |
21 |
3 |
9 |
0.22 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................GTACGTGTTGGTGGGTGTTTT........................................................................................... |
21 |
3 |
20 |
0.20 |
4 |
1 |
3 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGGTGGTGGGTGTTTTTGT........................................................................................ |
21 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGGTTGTGGGTGTTTTAG......................................................................................... |
20 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................TTTGAGGGTGTTATTGTGA...................................................................................... |
19 |
2 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGATGGTGGGTGTTTT........................................................................................... |
18 |
2 |
17 |
0.18 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................GCATTTATGTTTTTTTTTGGTTTCT................................................................................................................................. |
25 |
3 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGCTGGTGGGTGTTTTA.......................................................................................... |
20 |
3 |
18 |
0.11 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGTTGTTGGGTGTTTT........................................................................................... |
19 |
2 |
9 |
0.11 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................GAACGTGGTGGTGGGTGTTT............................................................................................ |
20 |
3 |
20 |
0.10 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTGGTGGGTGTTTTCGT........................................................................................ |
22 |
3 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................GAACGTGGTGGTGGGTGTTTT........................................................................................... |
21 |
3 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................CGTGATGGTGGGTGTTTTAG......................................................................................... |
20 |
3 |
16 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGATGGTGGGTGTTTTA.......................................................................................... |
20 |
3 |
19 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGGTGGTGGGTGTTTTCG......................................................................................... |
21 |
3 |
19 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................ACGTGTTGGTGGGTTTTT............................................................................................ |
18 |
2 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |