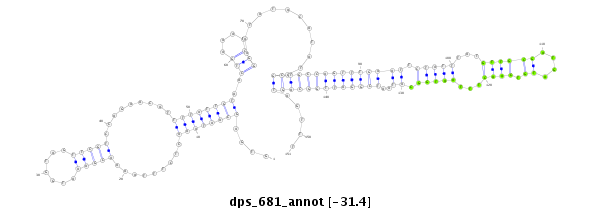
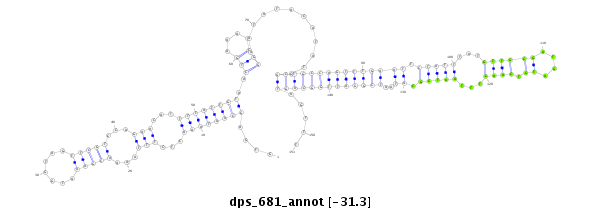
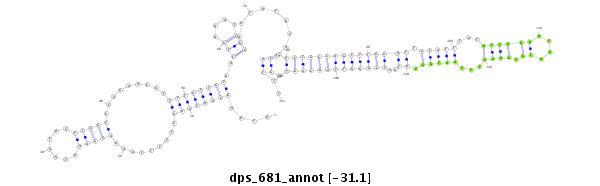
| .........GCAAAAACAAAAGACCAACT.............................................................................................................................................................................................................................. |
20 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................TAGTTAGTTGGTGAGGTAAA.................................... |
20 |
3 |
8 |
2.25 |
18 |
6 |
0 |
0 |
12 |
0 |
0 |
| .........................................................................................................................................................GTTGCGATGATGTCAACTTTAATGGC........................................................................ |
26 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .........................AACTCGAACCATGTCACGGA.............................................................................................................................................................................................................. |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...........................................................................................................................................................TGCGATGATGTCAACTTTAAT........................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................AACTTTAATGGCATAATCGGATTTGC.......................................................... |
26 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .............................CGAACCATGTCACGGACCAAGC........................................................................................................................................................................................................ |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................ATAATCGGATTGGCAGTAGTTT.................................................. |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................AAAAGACCAACTCGAACCATGT.................................................................................................................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .........GCAAAAACAAAAGACCAACTCGAAC......................................................................................................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................TGAATCCTATCTTTGTTGCGA........................................................................................... |
21 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......GAGCAAAAACAAAAGACCAAC............................................................................................................................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................................GCGATGATGTCAACTTTAATGGCAT...................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........AGCAAAAACAAAAGACCAACT.............................................................................................................................................................................................................................. |
21 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................ACTTTAATGGCATAATCGGATTGGC.......................................................... |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................GTAACACGTGGGAAACCT.... |
18 |
3 |
20 |
0.60 |
12 |
8 |
0 |
0 |
0 |
3 |
1 |
| ...............................................................................................................................................................ATGATGTCAACTTTAATTACAGA..................................................................... |
23 |
3 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................ATATGGTAGTTCGTCTAGAAG............................................................................................................................................................ |
21 |
3 |
7 |
0.14 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..............................................................................................................................AAGGCTGCTAGCTTGAATT.......................................................................................................... |
19 |
3 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................TAGTTAGTTGGTGAGGTAA..................................... |
19 |
3 |
20 |
0.10 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
| ......................................................................................................................................................................................................TTATCTGGGGAGGTAAAAAA................................. |
20 |
3 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .........................AACCCGAACCATGTGATGGA.............................................................................................................................................................................................................. |
20 |
3 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................ACTACATCTGGCTGCCAGTT................................................................................................................ |
20 |
3 |
13 |
0.08 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ........................................ACGGACCAAGGAGAGAAA................................................................................................................................................................................................. |
18 |
2 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |