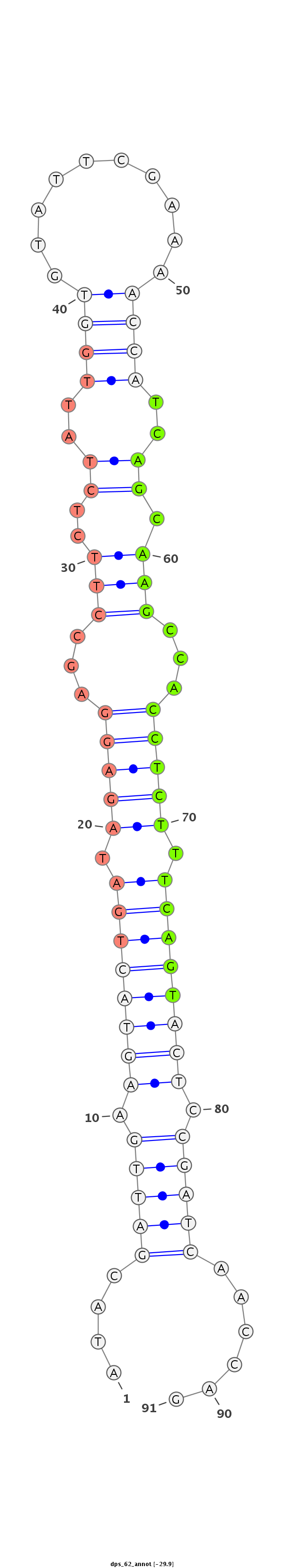
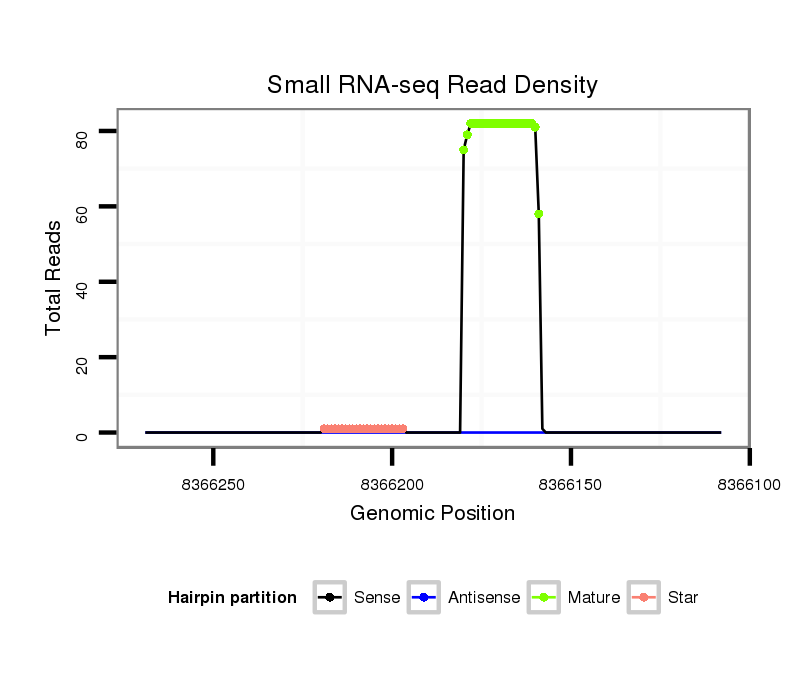
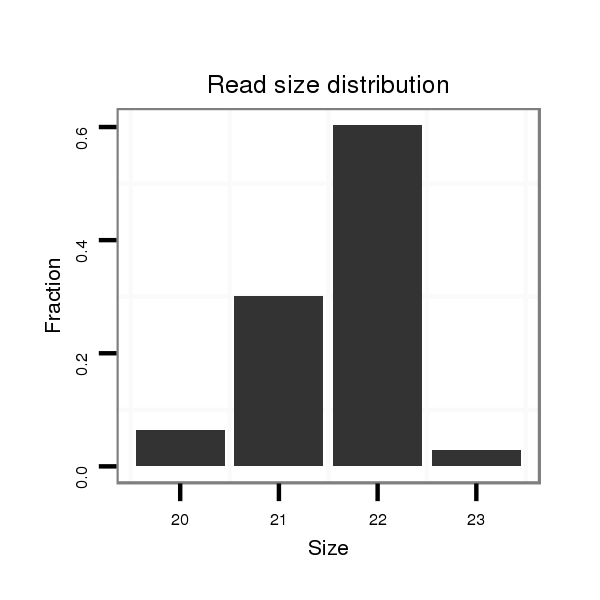
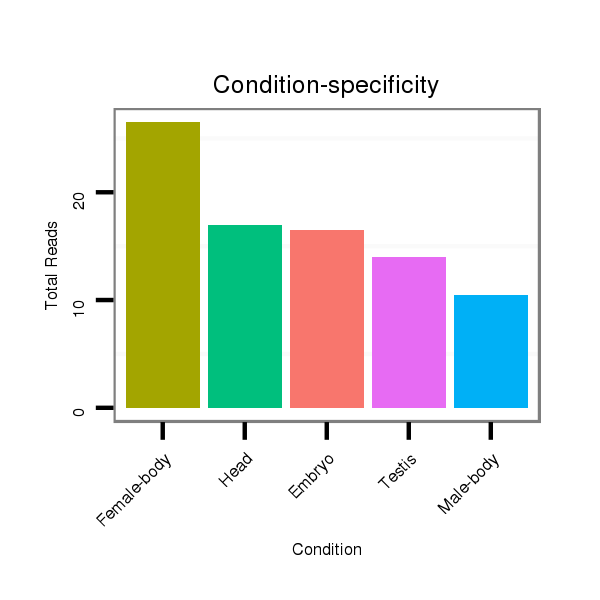
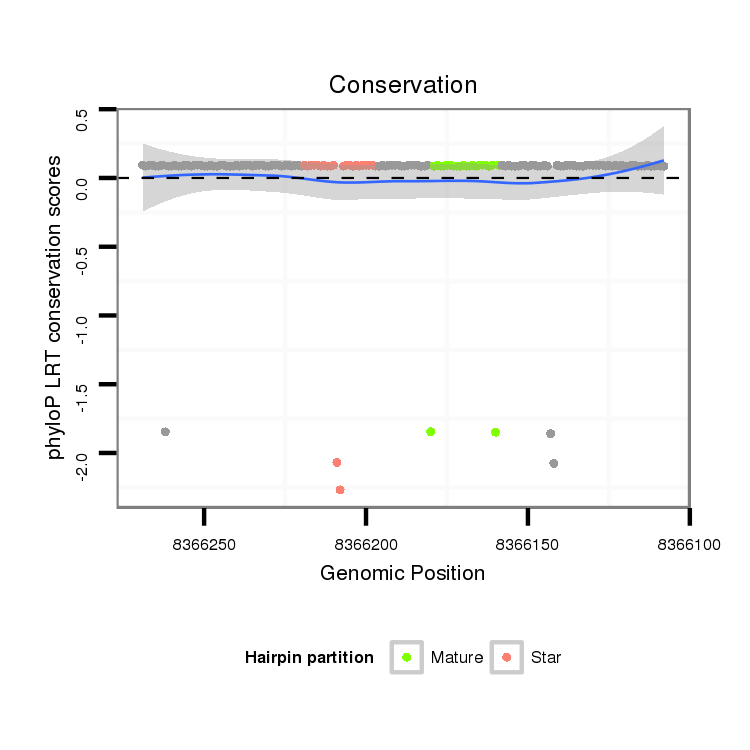
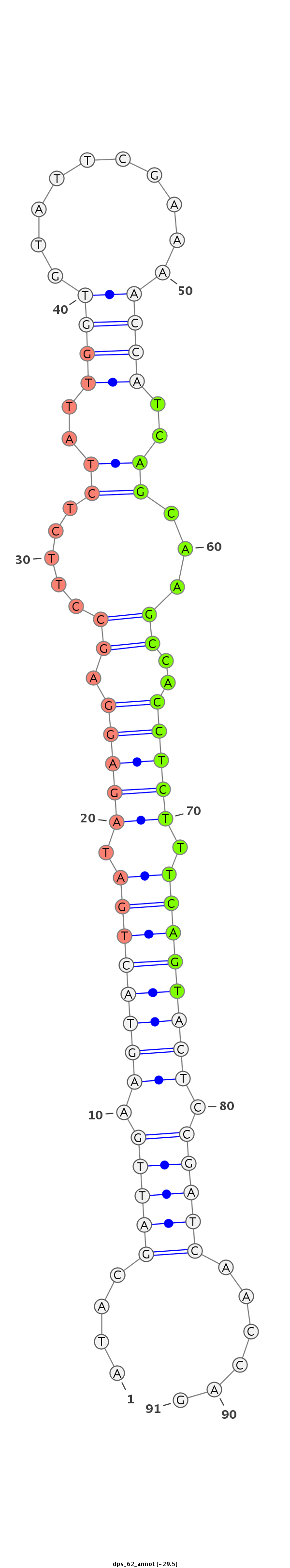
ID:dps_62 |
Coordinate:XL_group1e:8366158-8366219 - |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -29.5 | -29.2 | -29.2 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
AGATCGATCGACGATCTATGGAGGATCCCTGTTTCATACGATTGAAGTACTGATAGAGGAGCCTTCTCTATTGGTGTATTCGAAAACCATCAGCAAGCCACCTCTTTCAGTACTCCGATCAACCAGCATGGTATCCCAATATTCTGTGACGAACGCTGGAGG
***********************************....(((((.((((((((.(((((...(((..((..((((..........))))..)).)))...))))).)))))))).)))))......************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
GSM444067 head |
M059 embryo |
V112 male body |
GSM343916 embryo |
M062 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................TCAGCAAGCCACCTCTTTCAGT................................................... | 22 | 0 | 1 | 51.00 | 51 | 10 | 5 | 11 | 7 | 8 | 8 | 2 |
| .........................................................................................TCAGCAAGCCACCTCTTTCAG.................................................... | 21 | 0 | 1 | 22.00 | 22 | 12 | 6 | 2 | 1 | 1 | 0 | 0 |
| .........................................................................................TCAGCAAGCCACCTCTTTCAGC................................................... | 22 | 1 | 1 | 5.00 | 5 | 0 | 0 | 0 | 1 | 0 | 1 | 3 |
| .........................................................................................TCAGCAAGCCACCTCTTTCAGTT.................................................. | 23 | 1 | 1 | 4.00 | 4 | 1 | 2 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................CAGCAAGCCACCTCTTTCAGT................................................... | 21 | 0 | 1 | 3.00 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................AGCAAGCCACCTCTTTCAGT................................................... | 20 | 0 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGATAGAGGAGCCTTCTCTATTG......................................................................................... | 23 | 0 | 2 | 1.50 | 3 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| .........................................................................................TCAGCAAGCCACCTCTTTCA..................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................TCAGCAAGCCAGCTCTTTCAGT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGAGCAAGCCACCTCTTTCAGTT.................................................. | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................CTCAGCAAGCCACCTCTTTCAG.................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................TCAGCAAGCCACCTCTTTCGGT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................CAGCAAGCCACCTCTTTCAG.................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TCAGCAAGCCACCTCTTTCAGTA.................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TAAGCAAGCCACCTCTTTCAGT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................CTCAGCAAGCCACCTCTTTCAGTT.................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................TCAGCAAGCCACCTCTTTCAGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................TCAGCAAGCCACCTCTTTCAGTAA................................................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TTAGCAAGCCACCTCTTTCAGT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................CAATATACTGTGATGAACGCC..... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGAGGAGCCTTCTCTT............................................................................................ | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................AGATAGAGGAGCCTTCTCTATTG......................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TTAGAGAGGAGCCTTCTCTATTG......................................................................................... | 23 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TGATAGAGGAGCCTTCTCTAT........................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGATAGAGGAGCCTTCTCTA............................................................................................ | 20 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TAATAGAGGAGCCTTCTCTATTG......................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................TTTCAAACGATAGAAGCACTG.............................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................GCCATCTTTTTCAGTTCTCCG............................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................CTGCGACGAACGTCGGAGG | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
TCTAGCTAGCTGCTAGATACCTCCTAGGGACAAAGTATGCTAACTTCATGACTATCTCCTCGGAAGAGATAACCACATAAGCTTTTGGTAGTCGTTCGGTGGAGAAAGTCATGAGGCTAGTTGGTCGTACCATAGGGTTATAAGACACTGCTTGCGACCTCC
************************************....(((((.((((((((.(((((...(((..((..((((..........))))..)).)))...))))).)))))))).)))))......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM444067 head |
GSM343916 embryo |
V112 male body |
M040 female body |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................GGGGATAGTCATGAGGCTGGT......................................... | 21 | 3 | 4 | 1.25 | 5 | 4 | 1 | 0 | 0 |
| ....................................................................................................GGAGAAAGTCAAGAGGCTCGT......................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................CGGAAAAAGTCATGAGGCTGGT......................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................TGGAAAAAGTCATGAGGCCGGT......................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................GGTGATAGTCATGAGGCTCGT......................................... | 21 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................................................TGGAAAAAGTCAAGAGGCAAGT......................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .............................................................GGAAGAGATAACCACATAAGC................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................GGTGATAGTCATGAGGCTGGT......................................... | 21 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................GGAGATAGTCACGAGGCTAGT......................................... | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................GTGGAAAGTCACGAGGCTAGT......................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................GGGATAGTCATGAGGCTGGT......................................... | 20 | 3 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................GGAAAAAGTCAAGAGGCTAAT......................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................GGGATAGTCATGAGGCTCGT......................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................TGGTACCATAAGGATATAAG.................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................TTGTACCATACGGTTATAAT.................. | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................GTCGTACAATAGGGATCT..................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| dp5 | XL_group1e:8366108-8366269 - | dps_62 | confident | AGATCGATCGACGATCTATGGAGGATCCCTGTTTCATACGATTGAAGTACTGATAGAGGAGCCTTCTCTATTGGTGTATTCGAAAACCATCAGCAAGCCACCTCTTTCAGTACTCCGATCAACCAGCATGGTATCCCAATATTCTGTGACGAACGCTGGAGG |
| droPer2 | scaffold_13:198129-198290 + | dpe_138 | confident | AGATCGACCGACGATCTATGGAGGATCCCTGTTTCATACGATTGAAGTACTGATAGAGGATGCTTCTCTATTGGTGTATTCGAAAACCAACAGCAAGCCACCTCTTTCAATACTCCGATCAACCAGTCTGGTATCCCAATATTCTGTGACGAACGCTGGAGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 10/20/2015 at 07:08 PM