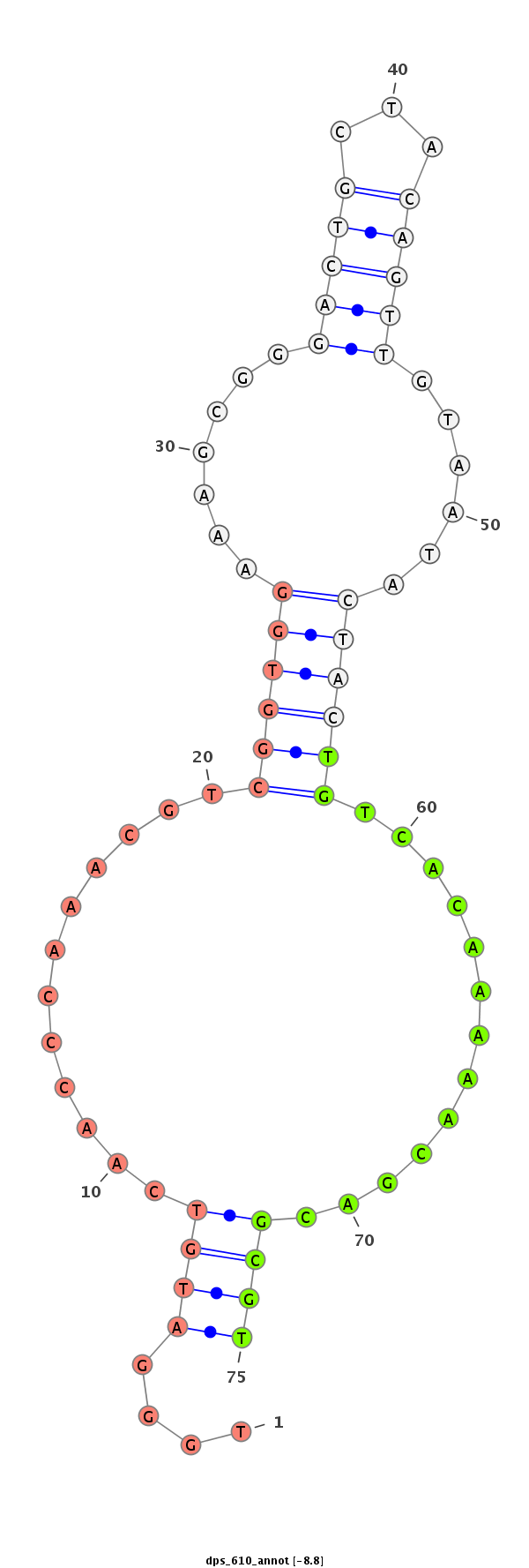
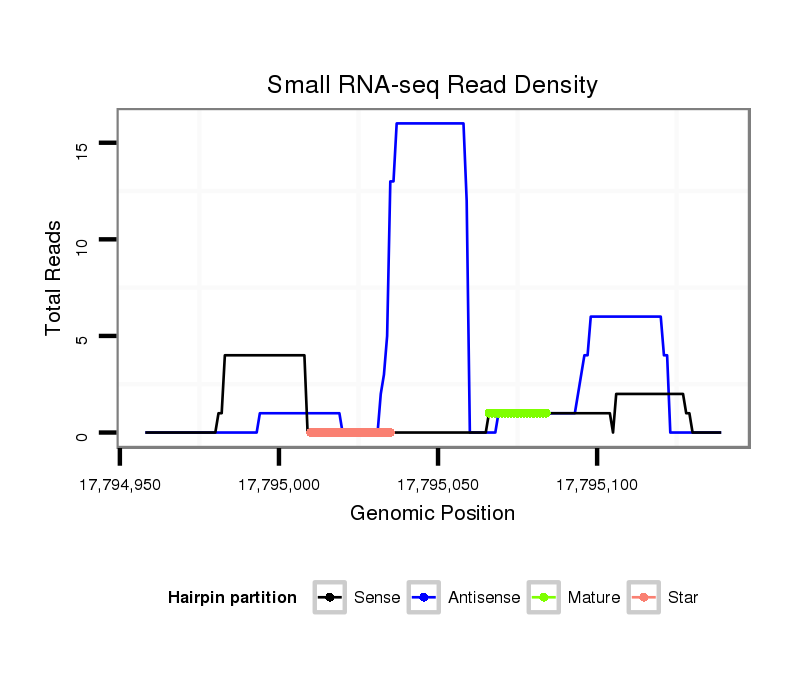

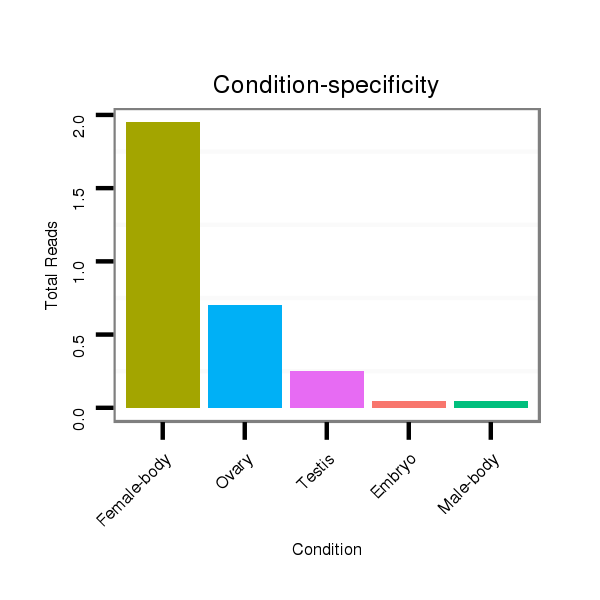
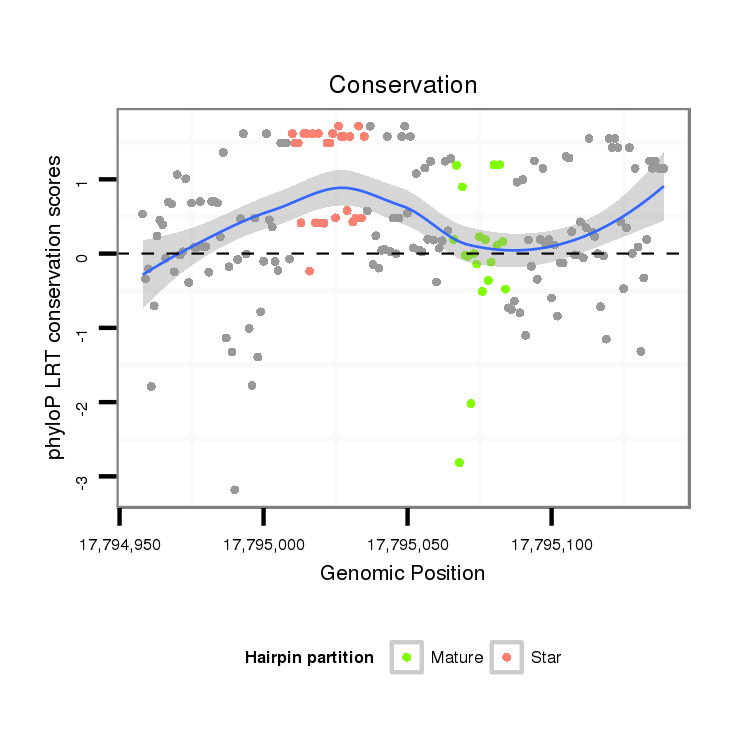
ID:dps_610 |
Coordinate:2:17795008-17795089 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.8 | -15.7 | -15.6 | -15.5 |
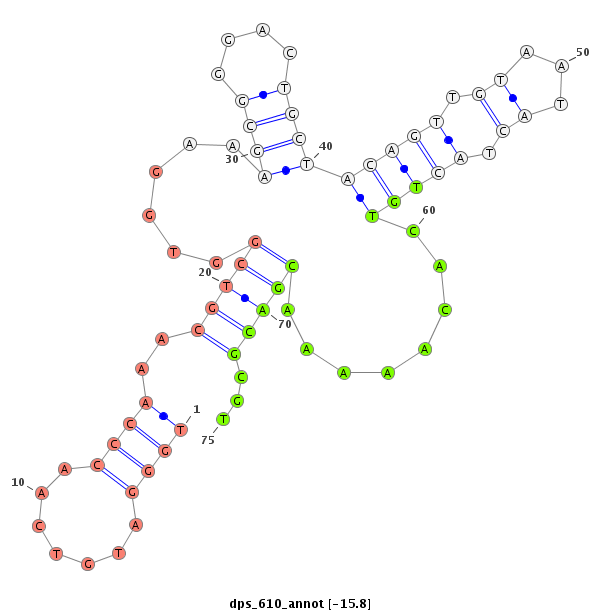 |
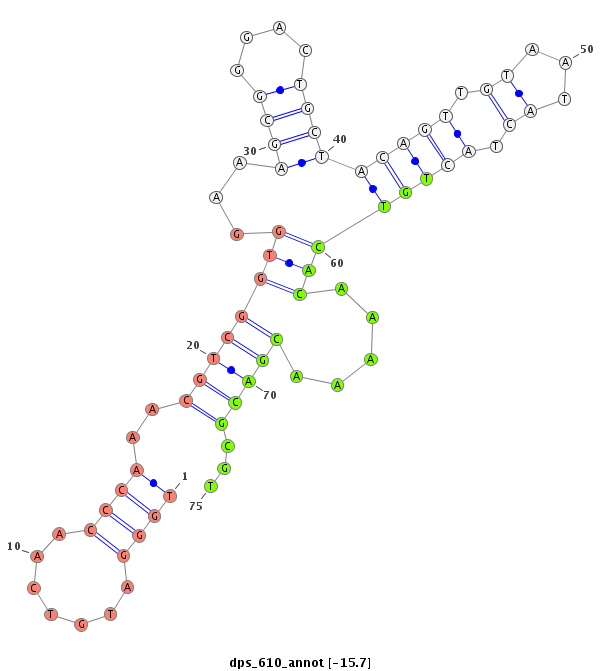 |
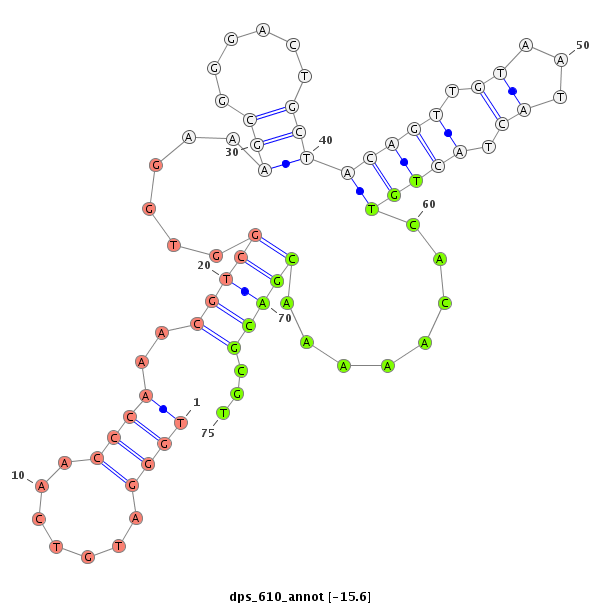 |
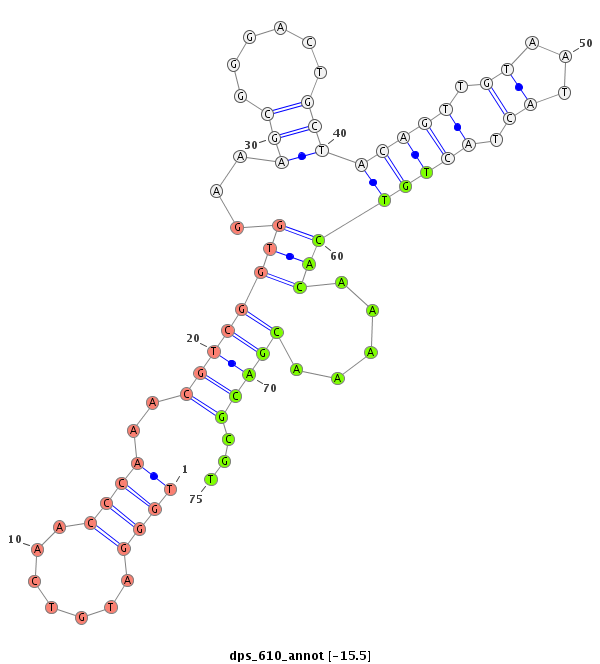 |
exon [Dpse\GA27310:1]; CDS [Dpse\GA27310-cds]; CDS [Dpse\GA27310-cds]; exon [Dpse\GA27310:2]; intron [Dpse\GA27310-in]
No Repeatable elements found
| ------------------------##########################----------------------------------------------------------------------------------################################################## CCGGTATGGACCTTCGGGCCAAATATGGAGGCTGAAAGTAGCAAGCACGGGTTGGGATGTCAACCCAAACGTCGGTGGAAAGCGGGACTGCTACAGTTGTAATACTACTGTCACAAAAACGACGCGTTGCAGGGTGATCCGCGCGGGTGCCGTTGTCGACGCAGACTGGGACCCGGAGAGCG ****************************************************....((((............((((((.......(((((...)))))......)))))).............))))******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
SRR902011 testis |
GSM343916 embryo |
M059 embryo |
SRR902012 CNS imaginal disc |
GSM444067 head |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................TGGAGGCTGAAAGTAGCAAGCACGGG................................................................................................................................... | 26 | 0 | 1 | 3.00 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TGTCACAAAAACGACGCGT....................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GGTCGAAACCGGGACTGCTGCA....................................................................................... | 22 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GCCGTTGTCGACGCAGACTGGGAC.......... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TATGGAGGCTGAAAGTAGCAAGCACGGG................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGCAGGGTGATCCGCGCGGG................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GCCGTTGTCGACGCAGACTGGG............ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TGGGATGTCAACCCAAACGTCGGTGG........................................................................................................ | 26 | 0 | 20 | 0.55 | 11 | 4 | 6 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TGATCCGAGCGGGTGCAGT............................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................ATCCGGGCGCGGGCCGTTGT.......................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................TTAGCAAGCACGGGTTGGGATG........................................................................................................................... | 22 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................TGTCAACCCAAACGTCGGTGGAAAGC................................................................................................... | 26 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TGGGATGTCAACCCAAACGTCGG........................................................................................................... | 23 | 0 | 20 | 0.25 | 5 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................CGGTCGAAACCGGGACTGCTG......................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGTCAACCCAAACGTCGGTG......................................................................................................... | 20 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TGGGATGTCAACCCAAACGTCG............................................................................................................ | 22 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................GGGATGTCAACCCAAACGTCGG........................................................................................................... | 22 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGCAGGGTGAACCGCGCGG.................................... | 19 | 1 | 13 | 0.15 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TTGGGATGTCAACCCAAACGTCGGTG......................................................................................................... | 26 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................TAAGACTAGTGTCAGAAAAA............................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................CGAAAACGAGACTGCTACAG...................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................ACTGACACAAAAACGACGCGTTG..................................................... | 23 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGTCAACCCAAACGTCGGTGGAAAG.................................................................................................... | 25 | 0 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................CGAAACCGGGACTGCTGCAG...................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................TGCAGGGTGAACCGCGCGGGTGCCG.............................. | 25 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGCAGGGTGAACCGCGCGGGTGT................................ | 23 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................GCAGGGTGAACCGCGCGGGTGC................................ | 22 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGCAGGGTGAACCGCGCGGGTA................................. | 22 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CGCGTTGCAGGGTGAACCGCGAGGA................................... | 25 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......TGGACCTTCGGGCCAAATATGGAGG....................................................................................................................................................... | 25 | 0 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .CGGTATGGACCTTCGGGC................................................................................................................................................................... | 18 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................CGGGGTGATCCGCGTGGGG.................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................GTCAACCCAAACGTCGGTGGA....................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................CAAGCACGGGTTGGGATGTCAAC...................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................AAGCACGGGTTGGGATGTCA........................................................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................CAAGCACGGGTTGGGATGTCAACC..................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGTCTACCCAAACGTCAGTG......................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................GTCAACCCAAACGTCGGTGGAAAGCG.................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GGCCATACCTGGAAGCCCGGTTTATACCTCCGACTTTCATCGTTCGTGCCCAACCCTACAGTTGGGTTTGCAGCCACCTTTCGCCCTGACGATGTCAACATTATGATGACAGTGTTTTTGCTGCGCAACGTCCCACTAGGCGCGCCCACGGCAACAGCTGCGTCTGACCCTGGGCCTCTCGC
*******************************************************....((((............((((((.......(((((...)))))......)))))).............))))**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
SRR902010 ovaries |
M059 embryo |
V112 male body |
M062 head |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................CTTTCGCCCTGACGATGTCAACATT................................................................................ | 25 | 0 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TTCGCCCTGACGATGTCAACATT................................................................................ | 23 | 0 | 1 | 3.00 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CGCGCCCACGGCAACAGCTGCGTCT................. | 25 | 0 | 5 | 2.80 | 14 | 13 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................CACCTTTCGCCCTGACGATGTCAACAT................................................................................. | 27 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GGCGCGCCCACGGCAACAGCTGCGTCT................. | 27 | 0 | 4 | 1.25 | 5 | 3 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................TCCGCCCTGACGATGTCAACATT................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AGGCGCGCCCACGGCAACAGCTGCGTCT................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................TCATCGTTCGTGCCCAACCCTACAGT........................................................................................................................ | 26 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TAGGCGCGCCCACGGCAACAGCTGCGT................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................CCTTTCGCCCTGACGATGTCAACATT................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTTTCGCCCTGACGATGTCAACAT................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................ACCTTTCGCCCTGACGATGTCAACATT................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................CCTTTCGCCCTGACGATGTCAACAT................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GGCGCGCCCACGGCAACAGCTGCGT................... | 25 | 0 | 4 | 1.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................GTGTTTTTGCTGCGCAACGTCCCACTA............................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CGCGCCCACGGCAACAGCTGCGT................... | 23 | 0 | 5 | 0.80 | 4 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................CGCCCACGGCAACAGCTGCGTCT................. | 23 | 0 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGCCATACCTGGAAGCCCGGTTTAT............................................................................................................................................................. | 25 | 0 | 14 | 0.57 | 8 | 4 | 0 | 2 | 1 | 0 | 0 | 1 |
| ..........................................................................................................................................GGCGCGCCCACGGCAACAGC........................ | 20 | 0 | 9 | 0.56 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................GCGCGCCCACGGCAACAGCTGCGT................... | 24 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................GCGCCCACGGCAACAGCTGCGTCT................. | 24 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CGCCCACGGCAACAGCTGCGT................... | 21 | 0 | 5 | 0.40 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................GCGCGCCCACGGCAACAGC........................ | 19 | 0 | 9 | 0.33 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AGGCGCGTCCACGGCAACAGCTGCGT................... | 26 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................GTTTTTGCTGCGCAACGTCCCACT............................................. | 24 | 0 | 12 | 0.25 | 3 | 2 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................GGCGCGCCCACGGCAACAGCTGCGTC.................. | 26 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TTGGCGCGCCCACGGCAACAGCTGCGT................... | 27 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AGGCGCGCCCACGGCAACAGCTACGT................... | 26 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................GGGTTTGCAGCCACCTTTC.................................................................................................... | 19 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................GCGCGCCCACGGCAACAGCT....................... | 20 | 0 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CGCGCCCACGGCAACAGCTTCGTCT................. | 25 | 1 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GGCGCGCCCACGGCAACAGCT....................... | 21 | 0 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................CTACAGTTGGGTTTGCAGCCACCTTT..................................................................................................... | 26 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............GCCCGGTTCATACCTCCGACT................................................................................................................................................... | 21 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CGCGCCCACAGCAACAGCTGCG.................... | 22 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................GTGTTTTTGCTGCGCAACGTCCCACT............................................. | 26 | 0 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......ACCTGGAAGCCCGGTTTATACCTCCGACT................................................................................................................................................... | 29 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................TCGTTCGTGCCCAACCCTACAGT........................................................................................................................ | 23 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........GGAAGCCCGGTTTATACCTCCGACT................................................................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CGCGCCCACGGCAACAGCTGCG.................... | 22 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CGCGCCCACGGCAACAGCTGCGTC.................. | 24 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................TGCGCCCACGGCAACAGCTGCGT................... | 23 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GCCCACGGCAACAGCTGCGTCT................. | 22 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TCGTGCCCAACCCTACAGT........................................................................................................................ | 19 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................GGTTTGCAGCCACCTTTCG................................................................................................... | 19 | 0 | 20 | 0.15 | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................GTTGGGTTTGCAGCCACCTTTCGC.................................................................................................. | 24 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGCCATACCTGGAAGCCCGGTT................................................................................................................................................................ | 22 | 0 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TTTGCTGCGCAACGTCCCACT............................................. | 21 | 0 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CCATACCTGGAAGCCCGGTTT............................................................................................................................................................... | 21 | 0 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TGGCGCGCCCACGGCAACAGCTTCGT................... | 26 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GGCGCGCCCACGGCAACAGCTTCGTCT................. | 27 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................GCGCGCCCACGGCAACAGCTTCGTC.................. | 25 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TGGCGCGCCCACGGCAACAGC........................ | 21 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TGGAAGCCCGGTTTATACCT......................................................................................................................................................... | 20 | 0 | 18 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................TGCGCGCCCACGGCAACAGC........................ | 20 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................ACGGCAACAGCTGCGTCTGA............... | 20 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................TTGGGTTTGCAGCCACCTT...................................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................GGTTTGCAGCCACCTTTC.................................................................................................... | 18 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................AGTTGGGTTTGCAGCCACCTTTC.................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................GTTCGTGCCCAACCCTACAGT........................................................................................................................ | 21 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GGCGCGCCCACGGCAACAG......................... | 19 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................CAGTTGGGTTTGCAGCCACCTTTCG................................................................................................... | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................GTGTTTTTGCTGCGCAACGTCCC................................................ | 23 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................GTCAACATCATGATGACTGTGTTTTTGCT............................................................ | 29 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TTGCGCGCCCACGGCAACAG......................... | 20 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GCAACGTCCCACTTGGCGCGCCCAC................................. | 25 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AACGGCAACAGCTGCGTCT................. | 19 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................GTTTTTGCTGCGCAACGTCCCAC.............................................. | 23 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................TGTTTTTGCTGCGCAACGTCCCACT............................................. | 25 | 0 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....TACCTGGAAGCCCGGTTTATACCTCC....................................................................................................................................................... | 26 | 0 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................CAACATCATGATGACTGTGTTT................................................................. | 22 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TTTGCTGCGCAACGTCCCATT............................................. | 21 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GCCATACCTGGAAGCCCGGTTTAT............................................................................................................................................................. | 24 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....TACCTGGAAGCCCGGTTTATACCT......................................................................................................................................................... | 24 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TTGCTGCGCAACGTCCCACT............................................. | 20 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGCCATACCTGGAAGCCCGGT................................................................................................................................................................. | 21 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CTGGAAGCCCGGTTTATACCT......................................................................................................................................................... | 21 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......ACCTGGAAGCCCGGTTTATAC........................................................................................................................................................... | 21 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AGCATAACCCTACAGTTGGGTTTGCAGCCACCTT...................................................................................................... | 34 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................GCCACCTTTCGCTCTGACGAT......................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................ACGTGCCCAACCCTACAGT........................................................................................................................ | 19 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CCTACAGTTGGGTTTGCAGGCACCT....................................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................ACCCTACAGTTGGGTTTGCAGCCAC......................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TGGGTTTGCAGCCACCTTTCTC.................................................................................................. | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................AGGGTTTGCAGCCACCTTT..................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................CCCTACAGTTGGGTTTGCAGCCACCTT...................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................CAGTTGGGTTTGCAGCCACCTTT..................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................GGTTTGCTGCCACCTTTCGCTCT............................................................................................... | 23 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................TTGTTCGTGCCCAACCCTACAGT........................................................................................................................ | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CCTACACTTGGGTTTGCAGCCAC......................................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................TCGTTCGTGCCCAACCCTACAG......................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:17794958-17795139 + | dps_610 | CCGGTATGGACCTTCGGGCCAAATATGGAGGCTGA-AAGTAGCAAGCACGGGTTGGGATGTCA--------ACCCAAACGTCGGTGGAAAGCGGGACTGCTACA----------------------------------------------------------------------------------GTTGTAATACTACTGTCACAAAAACGACGCGTTGCAGGGTGATCCGCGCGGGTGCCGTTGTCGACGCAGACTGGGACCCGGAGAGCG |
| droPer2 | scaffold_600:1519-1690 + | CCGGTATGGACCTTCGGGCCAAATATGAAGGCAGATAAGGAGCAAGCACGGGTTGGTATGTAA--------ACCCACACGTCGGTGGAAAGCGTGACTGCTACAACTG---------------------------------------------------------------------------------------------CCACTAAAACGACGCGTTGCAGGGTGAGCCGCGCGGGTGCCGTTGTCGACGCAGACTCGGACCTGGAGAGCG | |
| droWil2 | scf2_1100000004961:2378076-2378248 - | CAA---TGGACGTACG----------------GAAAACAAATCCAACTCGGGCTGGGATATCAGCCCTTCAGCCCAAACGTCTATAGAAGAAATGCCCACTGC-----------------------------------GTCTCG----CGGTCATCCCTCTGTT-------------GCCGTGATGGTTGTAGAACTACTG---CCAAACTTACGCGG---------------AGCAATGCTGAAGTCCTAGGAGACATGGG----------- | |
| droVir3 | scaffold_12963:5334425-5334500 + | TCGAAATG-------------------GATGTCAATATAGAAACAAATCGGGCTGGGATATCA--------GTCCAAACGTCGGTGGAAGAAGTGATCGCTAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000301666:44689-44828 + | CCACTCTG-------------------GAGAGTTTAAAGGATGAAACCAGGGATGGGATGTCA--------GCCCAAACGTCGGTGGAAGGCGTGACCGCTACCACCGACCGGCACGGTAGCGAG----------------------------------------------------------------------------------------------------CAATCCTCT------GCGTAGTCGATACAGACTCGGACCTTGAGAGCG | |
| droEle1 | scf7180000490212:15059-15281 - | CAAGTATG-------------------GAAGCGAAAAATAAATCAACCGGGGCTGGGATATCA--------GCCCAAACGACGGTGGAAGGCGTGTCTGCTACCACCGGCGGGCACGGGGAGGAGCAATCTTCTCTGCGTGTCGGCAGCGATCACACCTCTGAAATGAAGGGAGCAGGTCTTATTAGTTGTAACGCTTC-----------------------GGATAAGCCGCACAAATGCCATAGTCGAAGAAGACTCGGACCTAAAGAGCG | |
| droBia1 | scf7180000301698:29320-29404 - | CAGCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAATGCTACTGTCTT---GGCGTCTCAGGCAACGTTTAGTCGCACGGG-GCCGTTCTTGAAGTAGACTCCGGCTCGGAGAGCG | |
| droTak1 | scf7180000412366:385-582 - | CCAGT------------------------------------------------------------------------ACGTCGGTGGGAAGTACGACTGCTACCATCGGCGGGCACGGGGAGGAGCAATCCACTCTGCGTGTCGCCAGCGGTCGAACCCCTGGGATGGCGGGAGCAGGTCATTACAGCGGCAATGTTACTGCCAT---AACATCTCAGACAAAGTTAAGCCGTACGGGTGCCGTAGTTGTAGTAGACTCGGACTCGGAGAGCG | |
| droYak3 | v2_chrUn_4652:13841-14021 + | CCAAGAGTT--CTTTGTG--A--AATGGACAACAAAAACAGCTCAACCCGGGCTGGGATGTCA--------GCCCAAACGTCGGTGGAAAGCGTGACTGCTACCACGGG---------------------------------------CACGGAGCGC---------------------------------------ACGGGCACAGCAACGACTCAAGGGAGGCTAAGCCGCTCAGGTGCTATAGTCGATACAGACTTGGACCTGGAGAGCG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/17/2015 at 10:51 PM