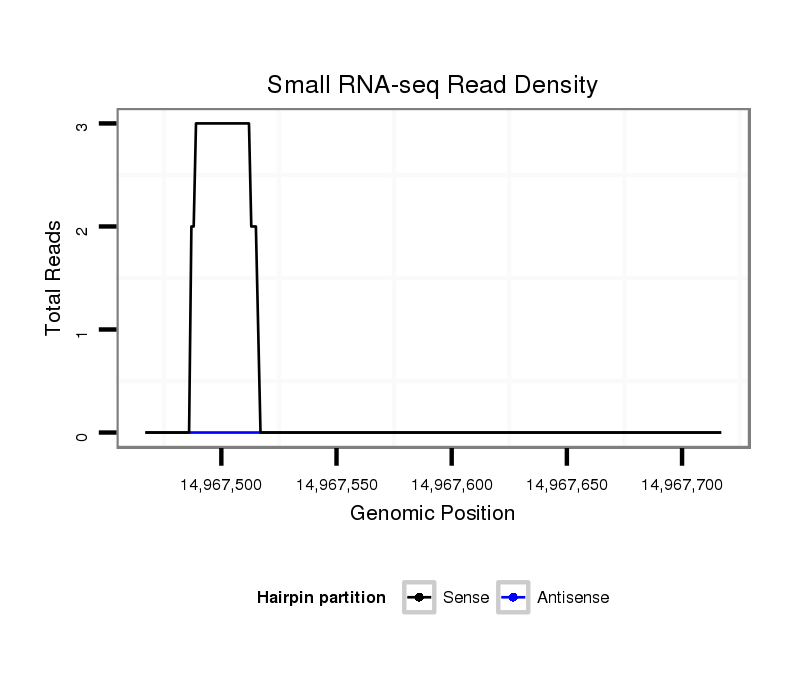
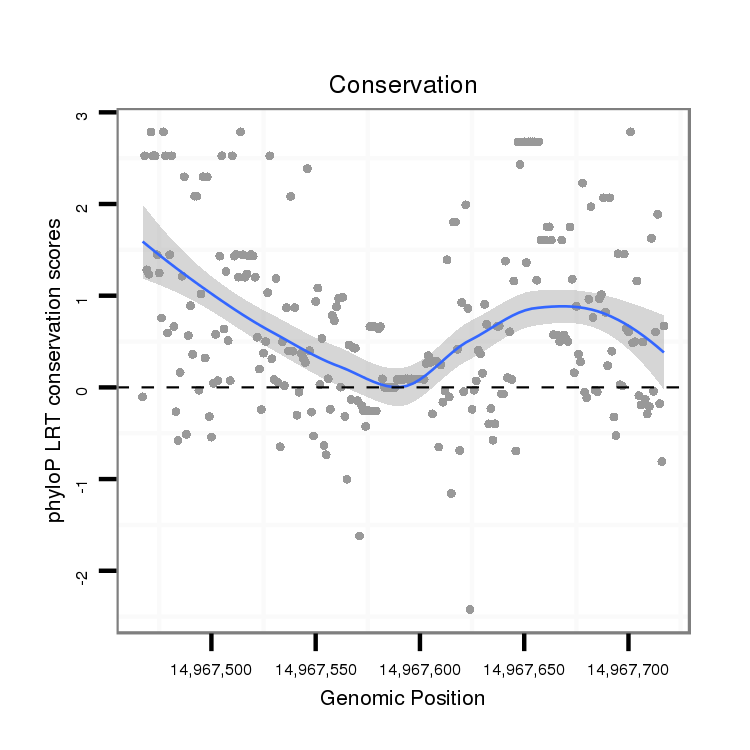
ID:dps_563 |
Coordinate:2:14967517-14967667 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [Dpse\GA15962:8]; CDS [Dpse\GA15962-cds]; exon [Dpse\GA15962:7]; CDS [Dpse\GA15962-cds]; intron [Dpse\GA15962-in]; intron [Dpse\GA15962-in]; intron [Dpse\GA15962-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGCCACCAATTCGTCCTCCTCTACGGCGTCGGCCCACTCCGGCGAAGAAGGTAAGCTAGTGCTCCAGCCGCCGGCAGACAGTTGCCATTTCGCTTGCCCACCCACACATACACCTACACCTACACCTACACTTAAGTCTAATCCCCAAACCGAATCCAAACCCTGTTCCTCCTCTGCTAATGTATATAACACGCTACTTTGCAACACTGCCACAACAGAAGCAGCAGCAACAGCAACAGCAACAAACATTA **************************************************..(((.(((((........((.(((((((((.......((((.((....................................................)).))))........))).......))))))...))........))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
V112 male body |
M022 male body |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTGC.......................................................................... | 19 | 2 | 1 | 214.00 | 214 | 214 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTGCT......................................................................... | 20 | 2 | 1 | 181.00 | 181 | 180 | 1 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTGCTA........................................................................ | 21 | 2 | 1 | 59.00 | 59 | 59 | 0 | 0 | 0 |
| .............................................................................................................................................................GAAGCCTGGTCCTCCTCTGC.......................................................................... | 20 | 3 | 15 | 46.47 | 697 | 697 | 0 | 0 | 0 |
| .............................................................................................................................................................GAAGCCTGGTCCTCCTCTGCT......................................................................... | 21 | 3 | 7 | 25.29 | 177 | 176 | 0 | 0 | 1 |
| ...............................................................................................................................................................AGCCTGGTCCTCCTCTGCTA........................................................................ | 20 | 2 | 1 | 19.00 | 19 | 19 | 0 | 0 | 0 |
| ...............................................................................................................................................................AGCCTGGTCCTCCTCTGCTAA....................................................................... | 21 | 2 | 1 | 13.00 | 13 | 13 | 0 | 0 | 0 |
| .............................................................................................................................................................GAAGCCTGGTCCTCCTCTG........................................................................... | 19 | 3 | 20 | 12.55 | 251 | 250 | 1 | 0 | 0 |
| ..................................................................................................................................................................CTGGTCCTCCTCTGCTAAACT.................................................................... | 21 | 3 | 4 | 8.75 | 35 | 35 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTGCTAA....................................................................... | 22 | 2 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGGTCCTCCTCTGCTAAACT.................................................................... | 20 | 3 | 8 | 5.50 | 44 | 44 | 0 | 0 | 0 |
| .................................................................................................................................................................CCTGGTCCTCCTCTGCTAAAC..................................................................... | 21 | 3 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 |
| ...............................................................................................................................................................AGCCTGGTCCTCCTCTGCT......................................................................... | 19 | 2 | 10 | 4.10 | 41 | 41 | 0 | 0 | 0 |
| .............................................................................................................................................................GAAGCCTGGTCCTCCTCTGCTA........................................................................ | 22 | 3 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| ...............................................................................................................................................................AGCCTGGTCCTCCTCTGCTAAA...................................................................... | 22 | 3 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTGCTT........................................................................ | 21 | 3 | 2 | 4.00 | 8 | 8 | 0 | 0 | 0 |
| .................................................................................................................................................................CCTGGTCCTCCTCTGCTAAA...................................................................... | 20 | 2 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTG........................................................................... | 18 | 2 | 8 | 3.88 | 31 | 31 | 0 | 0 | 0 |
| .................................................................................................................................................................CCTGGTCCTCCTCTGCTAA....................................................................... | 19 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| .............................................................................................................................................................GAAGCCTGGTCCTCCTCTGCTAA....................................................................... | 23 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGGTCCTCCTCTGCTAAA...................................................................... | 18 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ..................................................................................................................................................................CTGGTCCTCCTCTGCTAAA...................................................................... | 19 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ..................................................................................................................................................................CTGGTCCTCCTCTGCTAAAC..................................................................... | 20 | 3 | 6 | 2.17 | 13 | 13 | 0 | 0 | 0 |
| ................................................................................................................................................................GCCTGGTCCTCCTCTGCTAA....................................................................... | 20 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .................................................................................................................................................................CCTGGTCCTCCTCTGCTAAACT.................................................................... | 22 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................AGCCTGGTCCTCCTCTGC.......................................................................... | 18 | 2 | 19 | 1.42 | 27 | 27 | 0 | 0 | 0 |
| .............................................................................................................................................................CAAGCCTGGTCCTCCTCTGCT......................................................................... | 21 | 3 | 4 | 1.00 | 4 | 4 | 0 | 0 | 0 |
| ....................CTACGGCGTCGGCCCACTCCGGCGAA............................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................CGAAGCCTGGTCCTCCTCTGC.......................................................................... | 21 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ......................ACGGCGTCGGCCCACTCCGGCGAAGAA.......................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................CTGGTCCTCCTCTGCTAA....................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................GAACCCTGGTCCTCCTCTGCT......................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTTCTAA....................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................CTACGGCGTCGGCCCACTCCGGCGAAGAAG......................................................................................................................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................GCCTGGTCCTCCTCTGCTA........................................................................ | 19 | 2 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTGT.......................................................................... | 19 | 3 | 20 | 0.55 | 11 | 11 | 0 | 0 | 0 |
| .................................................................................................................................................................CCTGGTCCTCCTCTGCTA........................................................................ | 18 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................GACCTGGTCCTCCTCTGCTAA....................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................CCTGGTCCTCCTCTGCTAAG...................................................................... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGGTCCTCCTCTGCTAAAC..................................................................... | 19 | 3 | 16 | 0.44 | 7 | 7 | 0 | 0 | 0 |
| ..............................................................................................................................................................AGGCCTGGTCCTCCTCTGCTA........................................................................ | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................TGTTCCTCCTCTGCTAAACTG................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAGCCTGGTCCTCCTCTGCC......................................................................... | 20 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................AGCCTGGTCCTCCTCTGCTT........................................................................ | 20 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................................ACAGCCTGGTCCTCCTCTGCT......................................................................... | 21 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................GAAGCCTGATCCTCCTCTGC.......................................................................... | 20 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................GCCTGGTCCTCCTCTGCTAT....................................................................... | 20 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................CGAAGCCTGGTCCTCCTCT............................................................................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
CCGGTGGTTAAGCAGGAGGAGATGCCGCAGCCGGGTGAGGCCGCTTCTTCCATTCGATCACGAGGTCGGCGGCCGTCTGTCAACGGTAAAGCGAACGGGTGGGTGTGTATGTGGATGTGGATGTGGATGTGAATTCAGATTAGGGGTTTGGCTTAGGTTTGGGACAAGGAGGAGACGATTACATATATTGTGCGATGAAACGTTGTGACGGTGTTGTCTTCGTCGTCGTTGTCGTTGTCGTTGTTTGTAAT
**************************************************..(((.(((((........((.(((((((((.......((((.((....................................................)).))))........))).......))))))...))........))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M022 male body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................CGGACCAGGAGGAGACGATTT...................................................................... | 21 | 3 | 1 | 102.00 | 102 | 85 | 16 | 1 | 0 |
| .................................................................................................................................................................GGACCAGGAGGAGACGATTT...................................................................... | 20 | 2 | 1 | 63.00 | 63 | 50 | 13 | 0 | 0 |
| ................................................................................................................................................................CGGACCAGGAGGAGACGATT....................................................................... | 20 | 2 | 1 | 48.00 | 48 | 32 | 12 | 4 | 0 |
| ..............................................................................................................................................................TTCGGACCAGGAGGAGACGA......................................................................... | 20 | 2 | 1 | 38.00 | 38 | 26 | 12 | 0 | 0 |
| .................................................................................................................................................................GGACCAGGAGGAGACGATT....................................................................... | 19 | 1 | 1 | 38.00 | 38 | 32 | 6 | 0 | 0 |
| ..............................................................................................................................................................TTCGGACCAGGAGGAGACGAT........................................................................ | 21 | 2 | 1 | 34.00 | 34 | 25 | 7 | 2 | 0 |
| ...............................................................................................................................................................TCGGACCAGGAGGAGACGAT........................................................................ | 20 | 2 | 1 | 27.00 | 27 | 20 | 7 | 0 | 0 |
| ...............................................................................................................................................................TCGGACCAGGAGGAGACGATT....................................................................... | 21 | 2 | 1 | 24.00 | 24 | 18 | 6 | 0 | 0 |
| ................................................................................................................................................................TGGACCAGGAGGAGACGATT....................................................................... | 20 | 2 | 1 | 13.00 | 13 | 7 | 6 | 0 | 0 |
| ..................................................................................................................................................................GACCAGGAGGAGACGATTTGA.................................................................... | 21 | 3 | 4 | 10.25 | 41 | 33 | 8 | 0 | 0 |
| ..............................................................................................................................................................TTCGGACCAGGAGGAGACGATT....................................................................... | 22 | 2 | 1 | 9.00 | 9 | 6 | 2 | 1 | 0 |
| ..................................................................................................................................................................GACCAGGAGGAGACGATTT...................................................................... | 19 | 2 | 1 | 9.00 | 9 | 7 | 2 | 0 | 0 |
| .................................................................................................................................................................GGACCAGGAGGAGACGATTTG..................................................................... | 21 | 3 | 1 | 8.00 | 8 | 6 | 2 | 0 | 0 |
| ...................................................................................................................................................................ACCAGGAGGAGACGATTTGA.................................................................... | 20 | 3 | 8 | 7.00 | 56 | 38 | 16 | 2 | 0 |
| ..................................................................................................................................................................GACCAGGAGGAGACGATT....................................................................... | 18 | 1 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 |
| .............................................................................................................................................................CTTCGGACCAGGAGGAGACGA......................................................................... | 21 | 3 | 7 | 6.86 | 48 | 30 | 17 | 1 | 0 |
| .............................................................................................................................................................TTTCGGACCAGGAGGAGACGA......................................................................... | 21 | 2 | 1 | 5.00 | 5 | 3 | 2 | 0 | 0 |
| .................................................................................................................................................................GGACCAGGAGGAGACGAT........................................................................ | 18 | 1 | 2 | 4.00 | 8 | 8 | 0 | 0 | 0 |
| ................................................................................................................................................................CGGACCAGGAGGAGACGAT........................................................................ | 19 | 2 | 5 | 3.80 | 19 | 14 | 4 | 1 | 0 |
| .............................................................................................................................................................CTTCGGACCAGGAGGAGACGAT........................................................................ | 22 | 3 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 |
| ................................................................................................................................................................TGGACCAGGAGGAGACGAT........................................................................ | 19 | 2 | 4 | 2.50 | 10 | 6 | 4 | 0 | 0 |
| .................................................................................................................................................................GGACCAGGAGGAGACGATTTGA.................................................................... | 22 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................................TTTCGGACCAGGAGGAGACGAT........................................................................ | 22 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTGGACCAGGAGGAGACGAT........................................................................ | 20 | 2 | 2 | 1.50 | 3 | 1 | 2 | 0 | 0 |
| ...............................................................................................................................................................TCGGACCAGGAGGAGACGA......................................................................... | 19 | 2 | 10 | 1.50 | 15 | 13 | 2 | 0 | 0 |
| ...............................................................................................................................................................TTGGACCAGGAGGAGACGATT....................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................TGGACCAGGAGGAGACGATTT...................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................TTTGGACCAGGAGGAGACGAT........................................................................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................GACCAGGAGGAGACGATTTG..................................................................... | 20 | 3 | 6 | 1.00 | 6 | 6 | 0 | 0 | 0 |
| ..............................................................................................................................................................TTCGGACCAGGAGGAGACG.......................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................GACCAGGAGGAGACGATTTCT.................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................TTTCGGGCCAGGAGGAGACGAT........................................................................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................ATCGGACCAGGAGGAGACGATT....................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................GGACCAGGAGGAGACGATTG...................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................TCTCGGACCAGGAGGAGACGAT........................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................TGGACCAGGTGGAGACGATT....................................................................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..TGTGCTTAGGCAGGAGGAGAT.................................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................GGACCAGGAGGAGCCGATT....................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................GCACCAGGAGGAGACGATTT...................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................TTGGACCAGGAGGAGACGA......................................................................... | 19 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................TGACCAGGAGGAGACGATT....................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GTGTGTACGTGGTTGTGGATG................................................................................................................................ | 21 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................CTCGGACCAGGAGGAGACGAT........................................................................ | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................CTTCGGACCAGGAGGAGACG.......................................................................... | 20 | 3 | 15 | 0.20 | 3 | 3 | 0 | 0 | 0 |
| .............................................................GAGGGCGGAGGCCGTCTGTCG......................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................TGACCAGGAGGAGACGATTT...................................................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................TCGGACCAGGAGGAGACG.......................................................................... | 18 | 2 | 19 | 0.16 | 3 | 3 | 0 | 0 | 0 |
| ..............................................................................................................................................................TTCGGACCAGGAGGAGAC........................................................................... | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................ACCAGGAGGAGACGATTTG..................................................................... | 19 | 3 | 16 | 0.13 | 2 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................................................TTCAGACCAGGAGGAGACGA......................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................AAATCTAACGGGTGGGTTTG................................................................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................GTTCGGACCAGGAGGAGACG.......................................................................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................AATCTAACGGGTGGGTTTGT............................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................GTGTACGTGGTTGTGGATGT............................................................................................................................... | 20 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................ACGGGTGGGTGTGTACGA........................................................................................................................................... | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................TGGACCAGTAGGAGACGAT........................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................CTTCGGACCAGGAGGAGAC........................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:14967467-14967717 + | dps_563 | GGCCACCA---ATTCGTCCTCCTCTACG------------GCGTCGGCCCACTCCGGCGA------AGAAGGTAAGCTAGTGCTCCAGC-----------------------------------------CGCCGGC---AGACAGTTG--------------------------------------------------------------------------CCAT---TTCGCTTGCC---CACCCACACATACACCTACACCTACACCTACACTTAAGTCTAATCCCCAAACCGAATC-----------CA----------------------------AACCCTG------------TTCCTCCTC---------------------------------------------------------------------------------------TGCTAATGTATATAACACGCTACTTTGCAACACTGCCACAACAGAA---------GCA---------------------------GCAGC-------AACAG---------CAACAGCA---AC---------AAACA-TTA |
| droPer2 | scaffold_0:5442627-5442871 - | GGCCACCA---ATTCGTCCTCCTCTACG------------GCGTCGGCCCACTCCGGCGA------AGAAGGTAAGCTAGTGCTCCAGC-----------------------------------------CGCCGGC---AGACAGTTG--------------------------------------------------------------------------CCAT---TTCGCTTGCC---CACCCACACATACACCTA------CACCTACACTTAAGTCTAATCCCCAAACCGAATC-----------CA----------------------------AACCCTG------------TTCCTCCTC---------------------------------------------------------------------------------------TGCTAATGTATATAACACGCTACCTTGCAACACTGCCACAACAGAA---------GCA---------------------------GCAGC-------AACAG---------CAACAGAA---AC---------AAACA-TTA | |
| droWil2 | scf2_1100000004902:8232811-8233100 - | dwi_3145 | GGCCACCA---ATTCATC---ATCAT------------------CTGGAAATTCTGGGGA------AGAAGGTAAGCAGCAGCAGCAAGAT------GAAAATGAA--GAAGAGGAGGAGGAGGAGAAAG--------------AGAAGATGC---------------------------------------------AAAAAGATAATTTTAATGCCAATTGCAAT---TTTGCTTGTTCCTTGCCCAAA------------------------------TCC---------TCCAAATCT----CCCAATTGTCCCC---CTTG----------------TAT---------------C-------CC---------------------------------------------------------------------------------------TGCTAATGTAAATAACACGCTACTTTGCAACACTGCCACTACACACAATACAACAACAAAAACAACAACAGAAGCAACAACATCATCTAC------TATGTG---------TACTATTA----------------------- |
| droVir3 | scaffold_12723:4828096-4828350 + | TGCCACCGCGCATTCACC---CGCCTTG---GCG------GCGGCTGCTGTTGCCGCTGCCGCCTATCAGCTGTCCGCAA--CGTCAGCGGCGGCAGCAGC----------------------------------------AGCAGCGG--------------------------------------------------------------------------CCT-----------CCTCCTCCTCTGCG------------------------------TCC---------GCCACGGC-----------CGCTGCCTCTAGCTGCCAAG----CGGCCAGTGCGTT------------TGTGGCCTCGGCCAGCATGCTGGACATGCAACAACAACTGGAGGA----------------------------------------------------------------------------------------------GCA---------ACAGCAGCAGCAGCAGCAGCAACA------ACACC-------ACCAGCACTACCTGCA----------------------ACA-TCA | |
| droMoj3 | scaffold_6540:28869648-28869958 + | dmo_2106 | GGCCACCA---ACTCATC---ATCATCGACGGCGACGACGGCGTCAGTCAGTTCCGTGGA------AGAAGGTAAGGCAA--CAACGACAACAACAACAACAACGACACAACTACAACTACAACAACTACTGTCAGA---G--------------CAACAGCAGCTGCAACAGCAACGACAACAACAACAACAACAGCAACAACAACCATTAAATTCTAGCTGCAAT------------------AA-----------------------------------------------------------------------CTTATCCG----------------C--TTTGTTAGACCCAAAC-------CT---------------------------------------------------------------------------------------TGCCATTGTAAATAATACGCTACTACACTCTACTACCACATCAACA---------ACA---------------------------ACAGC-------AACAA---------CACCAACT---TTTAACGTCATGAACA-ACA |
| droGri2 | scaffold_15074:4004781-4005061 - | dgr_244 | GGCAACCA---ACTCATCCAGTTCGACA------------GCCTCTGCACACTCTACGGA------AGAAGGTAAGATGGAACAAGAACAACCACAACAACAACAA--GAACTAGAACAAGAACAACAAC--------------AGACGCTCGAACAACAAGCGCTACAACTG--------------------------------------------------CAAC---AAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------AACTGCAACAGCAACAGCAACTATTCAATGTTAACAATCATTCAAGCACAACGCCCCTAAATGCTATTGTAAATAACACGCTACTTTGCAACACTACCACAACAACA---------ACA---------------------------ACAAC-------TACAA---------AACAAGCT---TC---------AAATA-TTA |
| droAna3 | scaffold_12911:3204576-3204761 + | AGCCACCAATAGTTCCTC---C------------------------------TCCGGCGA------AGAAGGTAAGCCCGCGCAGCACCAG---------------------------------------CTTCATC---CAGA-------------------------------------------------------------------CAAA---------------ATGGGCTGCC---GAAGCGTA------------------------------TCC------CAAACCGTAAC-----------CCAAGTCCAAACCA----------------CGTCCTGTTC--------C-------TT---------------------------------------------------------------------------------------TGTCCCTGTAAATAACACGCTACTTTGCAACAC---------------------------------------------------------------TAAGAG---------CACCCTGCCACAT---------AACCA-TCC | |
| droBip1 | scf7180000396640:78428-78607 + | AGCCACCAACAGTTCCTC---C------------------------------TCCGGCGA------AGAAGGTAAGCCCGCGCAGCACCAG---------------------------------------CTTCATC---CAGA-------------------------------------------------------------------CCAA---------CCG---ATGGCCTGCC---AAACCGAG------------------------------TCC---------CCCGTAAC-----------CGTAACCCAAGTCCCAACCA----------AGCCCTGTTC--------C-------TT---------------------------------------------------------------------------------------TGTCCCTGTAAATAACACGCTACTTTGCAACAC---------------------------------------------------------------TAAGAG---------CACCCTGC-------------C--------- | |
| droKik1 | scf7180000302778:97710-97930 - | GGCCACCA---GTTCCTC---CTCATCG------------GCCTCGGGCCACTCCGGCGA------AGAAGGTAAGCCCGAGCAGCAAC-----------------------------------------CGTCAGCCGTCAGCAGCTG--------------------------------------------------------------------------CCCC---TTCGCTTGCC---CAACCAAG------------------------------ACCCAACCCG------AACCCAAAAC-----CACATCCTCAATCA----------------CGTCCCG------------------------------------------------------------------------------------------------------------CTCCCTTGTAAATAACACGCTACTTTGCAACACTGTCAGCACAGCA---------AC----------------------------TCTAA------CAGCAG---------CAGCCGCA---T------------CCA-AGA | |
| droFic1 | scf7180000453812:196174-196322 - | GGCCACCA---GTTCCTC---CTCGTCG------------GCATCGGGCCACTCCGGTGA------AGAAGGTAAGCCAGAGCAGCCAG-----------------------------------------CGACGGA---GCAGAGCTG--------------------------------------------------------------------------CCCC---ACCGCTCGCC---CACCCAAC------------------------------AC-----------------------------------------CC----------------CGTCCTG------------------------------------------------------------------------------------------------------------TCCCCCTGTAAATAACA---------------CCGA------------------------------------------------------------AAACAG---------CACCCTTA----------------------- | |
| droEle1 | scf7180000490994:598183-598383 + | GGCCACCA---GTTCCTC---CTCATCG------------GCATCGGGCCACTCCGGTGA------AGAAGGTAAGCCAGAGCAGCTCCGC--------------------------------------CCGTCAGC---CAGCAGCTG--------------------------------------------------------------------------CCCC---ACC---TCCG---TACCCGTATCCATATCT------------------------GTATCCT------TATCCACAGC-----CACATCCACATCCA----------------CATCCTG------------------------------------------------------------------------------------------------------------TTCCCTTGTAAATAACA---------------CTGCTATGA------------------------------------------------G------CAAGAG---------CACCCTTA------------ACCAGCA-TCA | |
| droRho1 | scf7180000779900:495745-495941 + | GGCCACCA---GTTCCTC---CTCATCG------------GCATCGGGCCACTCCGGTGA------AGAAGGTAAGCCAGAGCAGCATC-----------------------------------------CGTCAGC---CAGCAGCTG--------------------------------------------------------------------------CCCCACCTCCGCTTGCC---CACCCACA------------------------------ATC------CAGCCCGTATC-----------CCTATCCACATCCA----------------CATTCTG------------------------------------------------------------------------------------------------------------TTCCCTTGTAAATAACA---------------ATGCTAGCACAGCA---------ACG---------------------------TCTGCTACGAGCACGAG---------CACCC-------------------------- | |
| droBia1 | scf7180000302402:596712-596895 + | GGCCACCA---GTTCCTC---CTCATCG------------GCATCGGCTCACTCCGGTGA------AGAAGGTAAGCCAGAGCAGCTCC-----------------------------------------AGTCAGC---CAGCAACTG--------------------------------------------------------------------------CTCGAAAACCGCTTGCC---AACCTGAA------------------------------------------CCCGCATC-----------CATATCCCTGTCCGCAACCCTATC----CACATCCTG------------------------------------------------------------------------------------------------------------TATCCCTGTAAATAACA---------------CTG------------------------------------------------------A------CAAGAG---------CACCCTTA------------AT--------- | |
| droTak1 | scf7180000415256:338922-339085 + | CGCCACCA---GTTCCTC---CTCATCG------------GCATCGGGTCACTCCGGTGA------AGAAGGTAAGCCAGAGCAGCTCC-----------------------------------------CGTCAGGCGTCAGCACACC--------------------------------------------------------------------------TCCA---AT--------------------------------------------------------------------------------CGTTTCCCTATCCG----------------CATCCTG------------------------------------------------------------------------------------------------------------TATCCCTGTAAATAACA---------------CCGCCACCACTGAA---------ACG---------------------------TCTGC------CAAGAG---------CACCCTTA----------------------- | |
| droEug1 | scf7180000409804:1084392-1084590 + | AGCCACCA---GTTCCTC---CTCGTCA------------GCATCGGGCCACTCCGGTGA------AGAAGGTAAGCCAGAGCAGCTCC-----------------------------------------AGTCAGT---CAGCAGCTG--------------------------------------------------------------------------CCCC---ACCGCTTGCCAACCACCCACA------------------------------GTCAAACCACAA-----------------------TCCCTATCCG----------------CATCCTG------------------------------------------------------------------------------------------------------------TTTCCCTGTAAATAACA---------------CCGCTACCACTGAA---------ACG---------------------------TCTGT------TAAGAG---------CACCCTTA------------ACCACCATATA | |
| dm3 | chr3R:14844159-14844355 + | AGCCACCA---GTTCCTC---CTCATCG------------GCATCGGGTCAATCCGGTGA------AGAAGGTAAGCCAGAGCGGCTCC-----------------------------------------CGTCAGC---CAGCAGCCG--------------------------------------------------------------------------CCCC---TCAGCTTGCT---TACCCACA------------------------------ACTAAATCTCAATCCGAATC-----------CCTATCCTTATCTG----------------CATCCGG------------------------------------------------------------------------------------------------------------TTTCCCTGTAAATAACA---------------CTGCCACCACTGAA---------ACG---------------------------TCTGC------CAAGAG---------CACCCTTA----------------------- | |
| droSim2 | 3r:6413350-6413546 - | AGCCACCA---GTTCCTC---CTCATCG------------GCATCGGGTCAATCCGGTGA------AGAAGGTAAGCCAGAGCGGCTCC-----------------------------------------CGTCAGC---CAGCAGCCG--------------------------------------------------------------------------CCCC---TCAGCTTGCT---TACCCACA------------------------------ACTAAACCTCAATCCGAATC-----------CGTATCCTTATCTG----------------CATCCGG------------------------------------------------------------------------------------------------------------TTTCCCTGTAAATAACA---------------CTGCCACCACTGAA---------ACG---------------------------TCTGC------CAAGAG---------CACCCTTA----------------------- | |
| droSec2 | scaffold_77:93802-93998 - | AGCCACCA---GTTCCTC---CTCATCG------------GCATCGGGTCAATCCGGTGA------AGAAGGTAAGCCAGAGCGGCTCC-----------------------------------------CGTCAGC---CAGCAGCCG--------------------------------------------------------------------------CCCC---TCAGCTTGCT---TACCCACA------------------------------ACTAAACCTCAATCCGAATC-----------CGTATCCTTATCTG----------------CATCCGG------------------------------------------------------------------------------------------------------------TTTCCCTGTAAATAACA---------------CTGCCACCACTGAA---------ACG---------------------------TCTGC------CAAGAG---------CACCCTTA----------------------- | |
| droYak3 | 3R:3522194-3522392 + | AGTCACCA---GTTCCTC---CTCATCG------------GCATCGGGTCACTCCGGTGA------AGAAGGTAAGCCTGAGCGGCTCC-----------------------------------------CGTCAGC---CAGCAGCCG--------------------------------------------------------------------------CCCC---ACTGCTTTCC---TACCCACA------------------------------ACTAAACATCAGTCCGAATC-----------CGTATCCTTATCTG----------------CATCCAG------------------------------------------------------------------------------------------------------------TTTTCTTGTAAATAACA---------------CTGCCACCACTGAA---------ACG---------------------------TCTGT------CAAGAG---------CACCCTTA------------AT--------- | |
| droEre2 | scaffold_4770:6785219-6785421 - | AGCCACCA---GTTCCTC---CTCATCG------------GCATCGGGTCACTCCGGTGA------AGAAGGTAAGCCAGAGCGGCTCC-----------------------------------------CGTCAGC---CAGCAGCCG--------------------------------------------------------------------------CCCC---TCCTCTTGCT---TACCCACA------------------------------ACTAAATCTCAATCCGAATCCGAATC-----CGTATCCTTATCTG----------------CATCCGG------------------------------------------------------------------------------------------------------------TTTCCTTGTAAATAACA---------------CTGCCACCACTGAA---------ACG---------------------------TCTGC------CAAGAG---------CACCCTTA----------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:41 PM