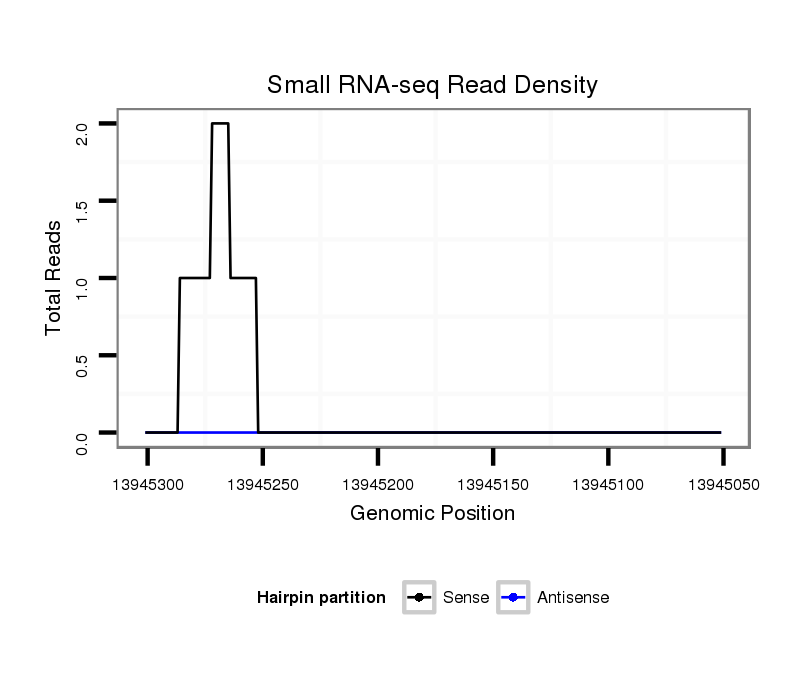
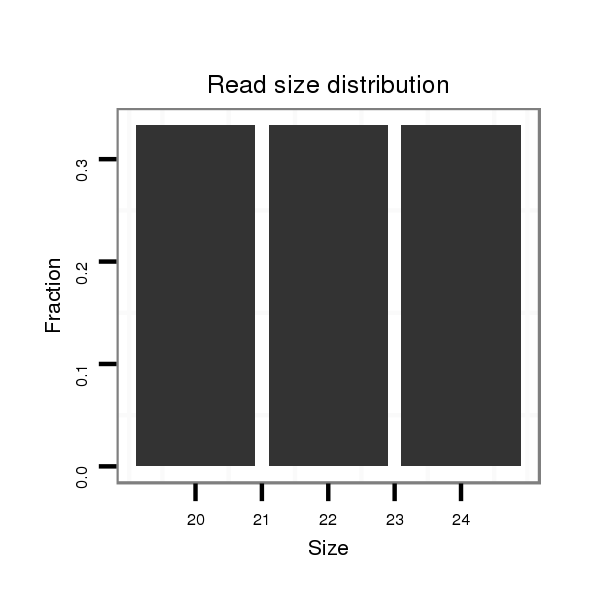
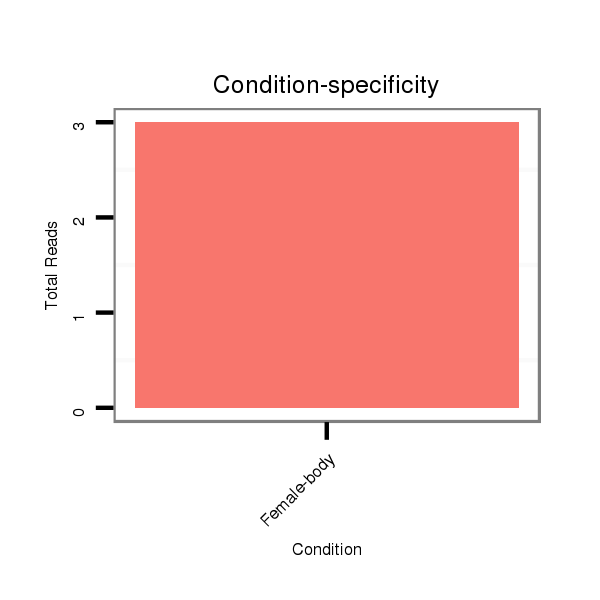
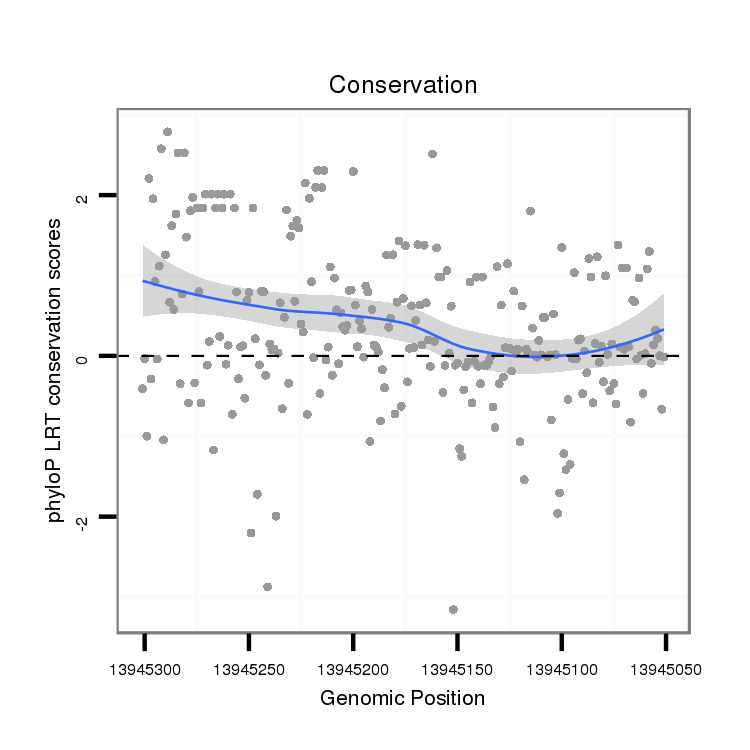
| dp5 |
2:13945132-13945382 - |
dps_533 |
AACA-ACA---AC----AACAGCAACCACATCATCTACAGCAACAACAACA---ACAACAGGCACCACAATTACACC------------------------AACAA----CA---ACAACA---GCAACAACAAAATGAAATGTCCAAA---------------------------------------------------------------------------TCCGCA---T------------TGG----GATTGCACTTTATCGAGGTAGATTCTATATGTATTCCACAA-----------ATCATAC----------------ATATT--------TAGT------GT--T------------------------------------TTTTTG-TGTGTGTGTG--------TTTTTCGTTGCACTACTAA--------CAGCA-CTA--------------------------------------------------------------------------------------------------------------------------CTGCTCC-C-------------------C-TCCCCTGCGTCTGCCGCTG---C-TCCCC |
| droPer2 |
scaffold_0:6498576-6498825 + |
|
AACA-ACA---AC----AACAGCAACCACATCATCTACAGCAACAACAACA---ACAACAACAGCAGGCACCACAAT------------------------TACA----CCAACAACAACA---GCAACAACAAAATGAAATGTCCAAA---------------------------------------------------------------------------TCCGCA---T------------TGG----GATTGCACTTTATCGAGGTAGATTCTATATGTATTCCACAA-----------ATCATAC----------------ATATT--------TAGT------GTATTTT--------------------------------------T-----------GTGTGTGTGTTTTTCGTTGCACTACTAA--------CAGCA-CTA--------------------------------------------------------------------------------------------------------------------------CTGCTCC-C----------------------CCCCTGCGTCTGCCGCTG---C-TCCCC |
| droWil2 |
scf2_1100000004909:5546401-5546630 + |
|
AAGACGCA---GC----AGCAGCAACAACATCAGCAGCAGCAGCAACAGCA---GCAGCAGCAGCAGCAACAACAAC------------------------AACA-GC--CACCTAC---------------------CAA------TGCCGCCA--TCTCAG--TC-----------------T------------------------------------------------A------------AGC----AATATCGATTTGTTCAGATATATAATAT------AGATA-----------------------------------------TATCTCTC----------TTGTTTC--------------------------------------T-----------GTTTCTTTGTTGGTCATTTAATGG-------CCTGGCATGG-C--------------------------------------------------------------------------AAATTT--------------------------TAGCTTC-------------------CCTC-------------------TCTCTGTCTCTGTCTCTG---TCTCTCT |
| droVir3 |
scaffold_12875:20356929-20357189 + |
|
TACA-GCA---GC----AATAGCAACAACAACAACAACAATATCAACAACA---GCAACTGCAACAACAATTATCAGTTCATCAACAACATCAACTACAACAACA-ACATCAACTACAACA---ACAACAACAACTCGA--------------------------ACAAGC---------------------------A----------------ACTGTTCTTTTTGGA---A------------TAG----TACTGCATTTTATTCCAATTGAATGTTT------AA--------------------------------------------------------TACGTGTTTTTTAC-------------------------------------------------GT-------------------------------------------GTCGCATTTAA--------------TGTTGTCTCA---------------------------------------------------------------------C---TGTCTC-----CCACTCTC---CCAC---------------TCTCTCTCTGCC------TCTGTGTC-TTCCC |
| droMoj3 |
scaffold_6680:15769711-15769890 + |
|
ACGA-ACA---AC----AACAACAACAACAACAACAGCAGCAACAGCAGCA---ACA---ACAGCCGCAATGACAACAGCAACAACAGT------------------------------------CAA-----GAAAACAATGCACAAA--------GCCAGGGC-------------------TAAAAG----------------------------------------------------------------------AGACATAGATATAGACAT-AT------ATAT-ATA--TATATATA----------------------------TATATATA-----------TATGTAA--------------------G----------------TG-TGTGTGT--GT------------------------------------------------GTGTGT--------------G--------------------------------------------------------------------------------------------------------------------------------------------------------TGT |
| droGri2 |
scaffold_10026:3242-3499 + |
|
CAA---CA---AC----AACAGCAACAACAGCAACAACAGCAGCAACAACA---ACAGCAGCAACAGCAA----------------------------------------------------------CAGCAACAAGAGCGACAAAAG---------------------------------------------------------------------------TCGACA---T------------CGA----CAATCAAAATCAACGCAATTAATGCTCA------TCCGGCCA--------------------------------------AAGTTGCA----CAGGGCTGTTATTC-------------------------------------------------AA-------------------------------------------TTTTC--ATAC--------------GTTTGCC--AGAGT-----------------TGCCAGGTCACATGTCAAATGTGGTGTGCGAGTCATGCTTACATTTGTA--------AGTCTCCTACTCCTCC-T-------------------CTTGCTCCGCT------TCTGCTGC-T-TCT |
| droAna3 |
scaffold_12943:4413849-4414141 - |
|
CAG---CATCAGC----AGCAA---CAGCAACAGCAGCAGCAGCAACAGCA---TCAGCAGCAGCAGCAATAGCAGCAGCAGCAACAGCAGCAGCAGCAATAGCA-GC----------------------------------------A--------GCAGCAAC-------------------AGCAGCAGCAACAGCAATAGCAGCATCTGCA---------GCAGCATCTTTAGCTTCAGCATCTGAGGCAGTTGCATTTT-------------------CCATTGCAGCT-----------TTCTTAC----------------CTGCT--------TTTT------TT--T------------------------------------TTTATA-TATTCCTTTT--------TTTTTTGTTAAAAAAGCAA--------CAGCAGCTG----C-----T--------------ACTCCACTTC---------------------------------------------------------------------GAGATACTTC---------------CCCACCT------------TC-------------CGCTTTTG---A-TTCTC |
| droBip1 |
scf7180000396708:996310-996533 + |
|
CAG---CA---AC----AACAA---CAACAA------------------------------------------------------------CA-------------------------ACAGCAACATCAACAAAATGAAATGTCCAAG---------------------------------------------------------------------------TCCGCT---T------------TGG----GACTGCATTTCATCGAGGTAGATTCTATAT--ATTCCACCAAAC------------------------------------AACTTATT----TTCGTCAT--TT-------------------TTGCTGCGCTTGTGT-CTTGTATCTTGT----GT--------CTTGCGTGT--------------------------GTTGCACATGTGTGTACTAGTAGTTG----------------------------------------------TATTGC--------------------------AAACTGCCCA---------------CTAACCA---------CAACT----------------AGCTG---CC-TTGC |
| droKik1 |
scf7180000302639:2653412-2653635 - |
|
CAC----------------CAA---CAACAT------------------------------------------------------------CAACAACAGCACCA-ACAACAATTACAACA---ACAGCAGCAGAACGAAATGTCCAAG---------------------------------------------------------------------------TCCGCT---T------------TGG----GACTGCACTTCATCGAGGTAGATTCTAT------TGTTTT----------------------------------------AACATATT----TTTGTCA------AACTTTTAACTTTAACTTTAAC----------------TG-TGTTTGTTTGT--------GTTGCGTTA---------------------------GTGCACATGT--------------TTTTGA-------------------------------------------------------------------------AAATTGCCCA---------------CTAACAAAGAAAAAAAAAACA----------------AGCTC---CCG---- |
| droFic1 |
scf7180000454073:3612353-3612542 - |
|
AGCA-A------C----ACCAA---CCACTA------------------------------------------------------------CA----------------------ACAACA---GCAGCAGCAGAATGAGATGTCCAAG---------------------------------------------------------------------------TCCGCG---T------------TGG----GACTGCATTTCATCGAGGTAGATTCTAT------TGCCCG----------------------------------------AACCTATT----TTCGTCTTGTG----------------------------------CCTTGTGG-CTTGTGTTTGT-------------------------------------------GCTGCACATA------CTCGTA--TGTTTGAG------------------------------------------------------------------------AAAATGCCCA---------------CTAACAA---------CAACA----------------TGCAA---CCGCTTC |
| droEle1 |
scf7180000491023:388095-388261 + |
|
AGCA-GCA---AC----AACAACAACAACAACAACAACAACAACAACAATA---ACAATAACAACAACAACAACAAC------------------------AACAA----CAACAACAGCA---ACCGCAACAAA----GGCAGCCAA---------------------------------------------------------------------------------------------------------TGCTGTAATTTATTAACTT----------------TGTGC----------------------------------------AAATT---GACT------C-------------------------------TCTTT----TTTTTG-TATTTATTTGT--------TTTTCATTG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droRho1 |
scf7180000780007:323710-323878 - |
|
AGCA-A------C----ACCAA---CCACAT------------------------------------------------------------CA-------------------------ACA---GCAGCAGCAGAATGAAATGTCCAAG---------------------------------------------------------------------------TCCGCT---T------------TGG----GACTGCATTTTATCGAGGTAGATTCTAT------TCGTCA----------------------------------------AACTTATT----TTTGTCTTGTGTTC-------------------------------------------------GT-------------------------------------------GTTGCACATGT--------------GTTTGAA--AAATA-----------------TGCC--------------------------------------------CAAATGCCCA---------------CTAACAA------------CA----------------TGCTA---C----GC |
| droBia1 |
scf7180000302075:2014771-2014968 - |
|
AGCA-GCT---AC----ACCAA---CCACAT------------------------------------------------------------CAACA-------------------GCAGCA---GCAGCAGCAGAATGAGATGTCCAAG---------------------------------------------------------------------------TCCGCA---T------------TGG----GACTGCATTTCATCGAGGTAGATTCTAT------TCCTCA----------------------------------------AACTTATT----TCTGTCTTGTGTTC-------------------------------------------------GT-------------------------------------------GCTGCACATGT--------------GTTTGTT--AGAAAAAAAAAAAAAAAGAAAG----------------AAAAGA--------------------------AAAATGCCCA---------------CTAA---------------CA----------------TGCTA---CCGCTTC |
| droTak1 |
scf7180000415700:302353-302500 + |
|
AACA-A-------------CAA---CCACAT------------------------------------------------------------CA-------------------------ACA------GCAGCAGAATGAAATGTCCAAG---------------------------------------------------------------------------TCCGCA---T------------TGG----GACTGCATTTCATCGAGGTAGATTCTAT------TCCTCAAA--------------------------------------AACCTATT----TCTGTCTTGTGTTC---------------------------------------------GTTCGT-------------------------------------------GCTGCACATGT--------------GTTTGAA---------------------------------------------------------------------------------A---------------CTAA---------------CA----------------TGCTA---CCA---- |
| droEug1 |
scf7180000409787:845308-845472 - |
|
AGCA-A------C----ACCAA---CCACAT------------------------------------------------------------CA-------------------------ACA---GCAGCAGCAGAATGAAATGTCCAAG---------------------------------------------------------------------------TCCGCT---T------------TGG----GGCTGCATTTCATCGAGGTAGATTCTAT------TCCTCA----------------------------------------AACTTATT----TTTGTCTTGTGTTT-------------------------------------------------GT-------------------------------------------GTTGCACATGT--------------GTTTGAA--A---------------------------------------------------------------------AGATTGCCCA---------------CTAACAA------------CA----------------TGCTA---CCGCTTC |
| dm3 |
chr3R:24736829-24736992 + |
|
AGCA-A------C----ACCAA---CCACAT------------------------------------------------------------CA-------------------------ACA------GCAGCAGAATGAAATGTCCAAG---------------------------------------------------------------------------TCCGCA---T------------TGG----GGCTGCACTTCATCGAGGTAGATTCTAT------TCCTCC----------------------------------------AATTTATT----TCTGTCTTGTTTTC-------------------------------------------------GT-------------------------------------------GCTGCACATGT--------------GTTTGAACCA---------------------------------------------------------------------AAAATGCCCA---------------CTAACAA------------CA----------------CGCTG---CCGCTTC |
| droSim2 |
3r:24098368-24098531 + |
|
AGCA-A------C----ACCAA---CCACAT------------------------------------------------------------CA-------------------------ACA------GCAGCAGAATGAAATGTCCAAG---------------------------------------------------------------------------TCCGCA---T------------TGG----GGCTGCACTTCATCGAGGTAGATTCTAT------TCCTCC----------------------------------------AATTTATT----TCTGTCTTGTTTTC-------------------------------------------------GT-------------------------------------------GCTGCACATGT--------------GTTTGAACCA---------------------------------------------------------------------AAAATGCCTA---------------CTAACAA------------CA----------------CGCTA---CCGCTTC |
| droSec2 |
scaffold_4:3600093-3600256 + |
|
AGCA-A------C----ACCAA---CCACAT------------------------------------------------------------CA-------------------------ACA------GCAGCAGAATGAAATGTCCAAG---------------------------------------------------------------------------TCCGCA---T------------TGG----GGCTGCACTTCATCGAGGTAGATTCTAT------TCCTCC----------------------------------------AATTTATT----TCTGTCTTGTTTTC-------------------------------------------------GT-------------------------------------------GCTGCACATGT--------------GTTTGAACCA---------------------------------------------------------------------AAAATGCCTA---------------CTAACAA------------CA----------------CGCTA---CCGCTTC |
| droYak3 |
3L:3535486-3535713 + |
|
CAG---CAGCAAC----AGCAA---CTGCAACAGCAGCAGCAGCAGCAAGATCGGCGGTGGCATCATTATCATGGCC------------------------ACGAT----CATCGACAAG---------------------------TGGCGAACAATTGAA---AAGTGCTTTTGCTTTGCTCTTGGGGGAACACGTG----------------CCGGGT---------------------------------------CTTTTTGGA-----TTTTTAT------TTCTTA----------------------------------------TACTTCTG--------TTTC------------------------------------------------------AA--------TTTTTTTTTGGACTGCTAT--------CG--------------------------------------ATCT---------------------------------------------------------------------TGAGTGCCGG---------------TGTACCT------------GT----------------CGCTG---CCGC-CA |
| droEre2 |
scaffold_4690:12275715-12275955 + |
|
AAAA-A--ACTAACTGCAACAACTACAACAT---CAACAATAACAACAACATCAACAGCAACAACAAGAACTACAAC------------------------A----------------ATA---ACAACAACAGCACGA--------------------------ACGCACA----------------------------------------------------CTCGCT---T------------TAA----CCT---------------CAAATTCTGTATGTACTGCTCAA-----------AACGCAAAAACCAAAATCGAGAAATATC--------TGTT------GTGTTTTT------------------------------------TTC-TAAACGTTAC--------CTTTTCCCCTCATCATTAAACGCTTGGAATCG-CTT--------------------------------------------------------------------------------------------------------------------------TTGCCCT-C-------------------T-CCC------------TCTA---A-G--TC |