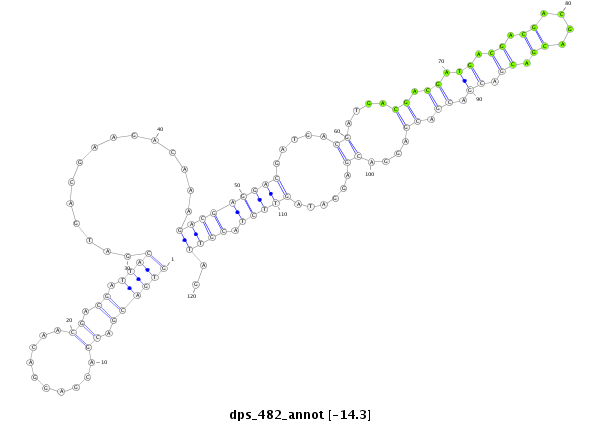
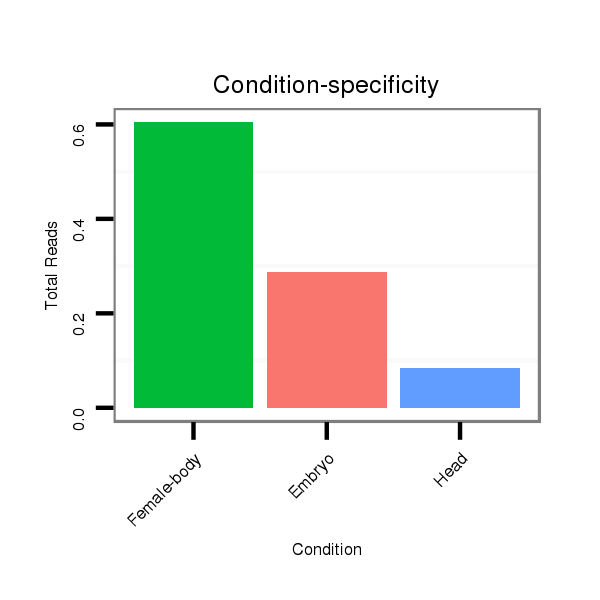
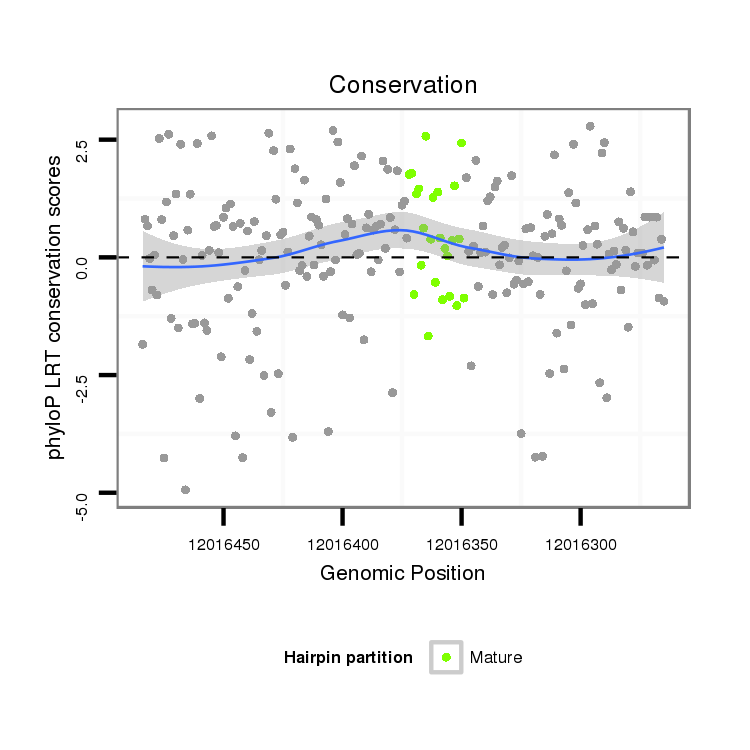
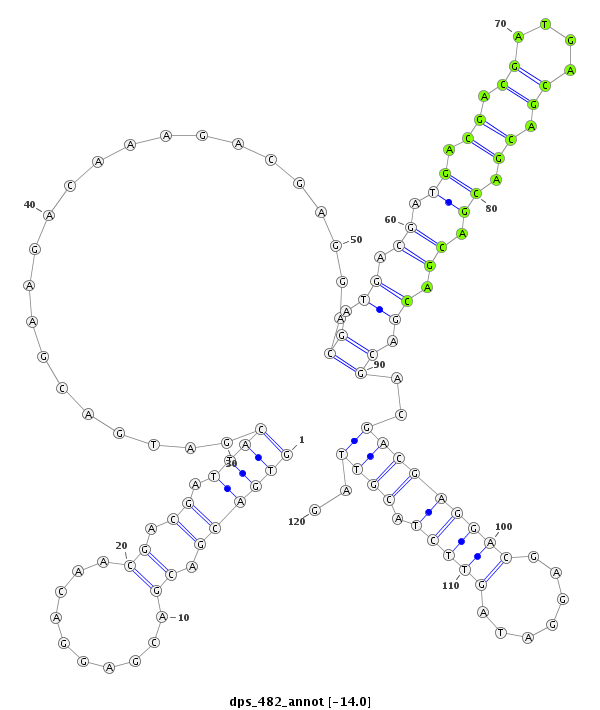
ID:dps_482 |
Coordinate:2:12016315-12016434 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -14.0 | -13.8 | -13.7 |
 |
 |
 |
CDS [Dpse\GA20512-cds]; exon [Dpse\GA20512:5]; exon [Dpse\GA20512:4]; CDS [Dpse\GA20512-cds]; intron [Dpse\GA20512-in]
| Name | Class | Family | Strand |
| (TCG)n | Simple_repeat | Simple_repeat | + |
| mature | star |
| ##################################################------------------------------------------------------------------------------------------------------------------------################################################## GGATATCGAGGAACAGGAAGACAAAGCGGAGAGGGACAAGGGCAAGAATGGTGACGACGACGAGGACAACGACGATTACGATGACGAAGACAAAGACGAGGACGATGACGATGACGACGATGACGACGACGACGACGACGACGACGAGGACGAGGATAGTTCTACGTTAGAAGATAGCGAAGATAGCGATGCCGACCCCAAAGTGGAGTTCCTGCTCGGC **************************************************((((((.((..........)).)).))))...............(((((((((.....((....((.((.((.((.((....)).)).)).)).))....))......))))).))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
SRR902010 ovaries |
M059 embryo |
GSM343916 embryo |
M062 head |
SRR902012 CNS imaginal disc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......GATGGAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 22 | 3 | 8 | 79.63 | 637 | 587 | 49 | 0 | 1 | 0 | 0 | 0 |
| ........ATGGAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 21 | 3 | 13 | 76.46 | 994 | 904 | 88 | 0 | 2 | 0 | 0 | 0 |
| ..........GGAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 19 | 2 | 8 | 7.75 | 62 | 61 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........ATGGAGAGGAAGACAAAGCG................................................................................................................................................................................................ | 20 | 3 | 20 | 5.85 | 117 | 106 | 11 | 0 | 0 | 0 | 0 | 0 |
| .......GATGGAGAGGAAGACAAAGCG................................................................................................................................................................................................ | 21 | 3 | 20 | 1.45 | 29 | 26 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...........GAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 18 | 2 | 20 | 1.15 | 23 | 23 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............AGAGGAAGACAAAGCGGG.............................................................................................................................................................................................. | 18 | 2 | 20 | 0.75 | 15 | 14 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AGTCCGACGATAGTTCTAC....................................................... | 19 | 3 | 6 | 0.67 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| ...............GGAAGACAAAGCGGGGTGG.......................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................GGCAAAATTGGTGACGACAAC............................................................................................................................................................... | 21 | 3 | 7 | 0.43 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........ATGGAGAGGAAGACAAAGCGGA.............................................................................................................................................................................................. | 22 | 3 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........TGAAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........GGAGAGGAAGACAAAGCGGA.............................................................................................................................................................................................. | 20 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............GGAAGACAAAGCGGGGTG........................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GAGGAAGACAAAGCGGGGTGG.......................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GACGACGATGACGACGACGACGAC.................................................................................... | 24 | 0 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GACGATGACGACGATGAC................................................................................................ | 18 | 0 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............AGAGGAAGACAAAGCGGATG............................................................................................................................................................................................ | 20 | 3 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................CAACGATGATTACTATGAGGA..................................................................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CGACGACGACGACGACGAC........................................................................... | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................GGCAAAATTGGTGACGAC.................................................................................................................................................................. | 18 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................CAAAGTGGAGTATCTGCTC... | 19 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................TGACGATGACGAGGATGACGA.............................................................................................. | 21 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................CGACGACGACGAGGACGAGGA................................................................ | 21 | 0 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........ACGGAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 21 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................ACGATGAGGACGATGACGACGACGATG...................................................................................... | 27 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................CGACGATGACGACGACGACGACGACG................................................................................ | 26 | 0 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......AGAGGGAGAGGAAGACAAAGCGG............................................................................................................................................................................................... | 23 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................GACGACGACGAGGACGAGGA................................................................ | 20 | 0 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................GAGGACGATGACGATGACT........................................................................................................ | 19 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................ACGACGACGAGGACGAGGA................................................................ | 19 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GACGACGATGACGACGACGAC....................................................................................... | 21 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TGACGACGAAGACGCCGATGACGAC.................................................................................... | 25 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................CGACGACGACGACGACGAGGA...................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ACGACGATGACGACGACG......................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............AGGAAGACAAAGCGGGAA............................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GACGACGATGACGACGACGA........................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................GATGAGGACGATGACGATGAC......................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................ATGAGGACGATGACGATGAC......................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TGACGACGACGACGACGAC................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................GACGACGACGACGACGAGG....................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................GATGACGACGACGACGACGACGAC.............................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CCTATAGCTCCTTGTCCTTCTGTTTCGCCTCTCCCTGTTCCCGTTCTTACCACTGCTGCTGCTCCTGTTGCTGCTAATGCTACTGCTTCTGTTTCTGCTCCTGCTACTGCTACTGCTGCTACTGCTGCTGCTGCTGCTGCTGCTGCTCCTGCTCCTATCAAGATGCAATCTTCTATCGCTTCTATCGCTACGGCTGGGGTTTCACCTCAAGGACGAGCCG
**************************************************((((((.((..........)).)).))))...............(((((((((.....((....((.((.((.((.((....)).)).)).)).))....))......))))).))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
V112 male body |
|---|---|---|---|---|---|---|---|---|
| ........TACCTCTCCTTCTGTTTCGCC............................................................................................................................................................................................... | 21 | 3 | 13 | 0.62 | 8 | 8 | 0 | 0 |
| ........TACCTCTCCTTCTGTTTCGC................................................................................................................................................................................................ | 20 | 3 | 20 | 0.25 | 5 | 5 | 0 | 0 |
| .......CTACCTCTCCTTCTGTTTCGC................................................................................................................................................................................................ | 21 | 3 | 20 | 0.25 | 5 | 5 | 0 | 0 |
| ...........................................................TTGTCCTGTTGCTGCTAAT.............................................................................................................................................. | 19 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ...........................................................................................................................GCTGCTGCTGCTGCTGCTG.............................................................................. | 19 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| ...........................................................TTGTCCTGTTGCTGCTAA............................................................................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 |
| .........CCCTCTCCTTCTGTTTCGCCC.............................................................................................................................................................................................. | 21 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 |
| .......................................................................TGGAAATGCTACTGCTTGT.................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ....................................................................................................................................GCTGCTGCTGCTGCTC........................................................................ | 16 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:12016265-12016484 - | dps_482 | GGAT-------------------ATCGAGGAAC---------------AGGAAGACAAAGC---GGAGA--------GGGAC---AAGGGCAAGA------ATGGTGACGACGAC------------------GAGGAC------------------------AACGACGA------------TTA---------CGATG---------------AC---------------------GAAGACAAAGACGAGGACGAT------------------------------GA------------CGATGACGACG-------------ATG---ACGACGACGA---------------------------------------------------------------------------CGAC------GACGACGACGAG------GA---CGAGGATAGTTCTACGTTAGAA--GATA--GCGAAGATA------------GCGA------TGCCGACCCCAAAGTGG---------------AGTTCCTGCTC----------------------------GGC |
| droPer2 | scaffold_0:8482837-8483041 + | GGAT-------------------ATCGAGGAAC---------------AGGAAGACGAAGC---GGAGA--------GAGAC------AAGGGCA------AGAATGATGACGAT------------------------------------------------------------------------------------GACG------------------------------------AAGACAAAGACGAGGACGAT------------------------------GACGATGACGACGAT------GACGACGATGACG-ACGATGACGACGATGACGACGACGA---------------------------------------------------------------------CGA------------------------------CGAGGAAAGTA---CGCCAGAA--GATA--GCGAAGATA------------GCGA------TGCCGACCCCAAAATGG---------------AGTTCCTGCTC----------------------------GGC | |
| droWil2 | scf2_1100000004558:188870-189082 + | TGAC-------------------GAAGAGGATGAAAATGATA------ATGGGGA---------AGA--T---------GAT---GAAGAAGATA------ATGACGAAGACAACGACAAAGACAATGAC---GA------------------------------------------------TG---------ATAATGATGAA---G---------------------------------ACAATGATGAAGACGAT---------------GATGAAGACAATGAT---------------------GAAGAAG---ATAA-AGAAAA------------------------------------------------------------------------------TGACGACGATGAC------GGGGATAATGATGACGA---------------TG---ATGAGA-----ATG--ACAAAGATG------------ACGA------CGAAGA-----------------CGATGAAGA---------------------------------------AGA | |
| droVir3 | scaffold_12875:15517071-15517274 - | TGAC-------------------AATGACGAGG---------------ACGACGATGAAAA---CGA--T---------------GAAGACAATG------ACGATGAAGATGAG------------------GACG---------------------------AAA----ATG--------------------ATGATGATGAA---GACAATGAT---------------------GAAGATGAGGATGAAAATGGT------------------------------GA------------CGATGACGAAG-------------ACG---------A---------------------------------------------------------CGATGAGAA---CGACAACGACAAC------------------------GATAAAGATAATGACG---ATGACGAA--GACG--ATGAAGACA------------ACGA------CGACGAAGACGAAGA--------------------------------------------------------GGA | |
| droMoj3 | scaffold_6359:1107463-1107665 - | G-AT-------------------ATGGATGAGG---------------AAGCCGGAGAAGG---CGA--T------GGGGATGATGACAATG---------ATGACGATGATGCC---AATGATGACGATGA---------------------------------------------------TGATGATGATGACGATGATGAT---GAT------G---------------------------ACGA---------------------------------------------------------------------TGGTG-ATGATGATGACGATGGTGACGATGATGACGAGGAAGACGACGAAGATGA---------------------------------------------------------------------------------------TG---ACG-------------AAAATGA------------TGACGATGATGTCGA--------------CATTGATGACTGTCC---------------------------------------AGC | |
| droGri2 | scaffold_15245:12322255-12322449 - | TGGC-------------------GATGACGATG---------------ACGAGGACGATAC---TGA--C---------------GACGATAACG------ATG---AC------------------AATGATAATG---------------------------------------------------ATGACGAAAATGACGAT---GAT------GAGGAC---------------GAAGACGAT------GACGAG------------------------AATGGAGA------------CGATGATGAAG-------------ATG---------AAGA------------------------------------------------------------------CGAAGA---CGAA------GACGATGA---------------CGATGACGAGA---ATGGAGAA--GATG--ATGAAGACG------------AAGA------CGATGA-----------------CGAGAATGT---------------------------------------AGA | |
| droAna3 | scaffold_12088:3374-3585 - | G-AT-------------------GACGATGACG---------------ATGACGATGACGA---TGA--C---------GAT---GACGATGACG------ATGACGATGACGAT------------------GACG---------------------------------------------------------ATGACGATGACGATGACGATGAC---------------------GATGACGAT------GACGAT------------------------GACGATGA------------CGATGACGATGACG---ATGA-CGATGA------------------------------------------------------------------------------CGATGACGATGAC------------------------GATGACGATGACGATG---ACGATGAC--GATG--ACGATGACG------------ATGA------CGATGACGATGACGATG---------------A----CTGCCC----------------------------GCC | |
| droBip1 | scf7180000396735:1662158-1662351 + | G-AT-------------------GACGAGGAAG---------------ATGAGGATAATGAAGACGA--T---------GAT---GAAGAAGATA------ATGGCAATGATGAC------------------G------------------------------------------------------------AAGACGAAGATGAT---------A---------------------------ACGATGGGGACGAA---------------GATAACGAGGATGAC------------------GAT---GAAG---ATCA-GGACGA------------------------------------------------------------------------------CAATAATGACGAC------GAGAACGATGAGGACGG---------------GG---ATGAGGAA--GACG--ATGAAGGCG------------ACGA------AGATGA--------------------AGATAA---------------------------------------TGA | |
| droKik1 | scf7180000301810:6336-6566 - | ATCTCAAGGAACTGGGACCAGAGGAGGAG--------------------------------------------------CAGGAGGAGCAGA---------AGGAGGACGAAGGC------------------GAAGAC------------------------GAAG----ATGACTACGATG--------ATGAGGAGGAGGAGGAG---------G---------------------------ATTTG-------------------------------------------------------------------------------------------------------------------GGAGACGGTGCTGATGAAGAGGGCGAGGAGGAGGAGGAGTC---CTACGACGACTA------------------------------CGAGCAGTCCTCGAGGACAGA---------------------------------AGAACGCCGC--------------CGCCGCCG-------T----CCGCCCGTTCAAAGCGCCTTAAGTGTCCGGAGTAGGC | |
| droFic1 | scf7180000453955:681993-682209 + | AGAT-------------------AACGACGAAG---------------ATGAAGACGAAGA---TGA--A---------------GGAGATGATA------ACGATAAGGGTGAC------------------G------------------------------------------------------ACGATGAGAATGATGAC---GAA------GAAAATGACAATGACGATGAGGAAGATGAAGACGAAGAAGAT------------------------------AA------------------------TG---ATAA-GGATGA------------TGAGGATGAGGATGAGGATAATGAGGATGG---------------------------------------------------------------------------------------GG---ATGGAG-----ACG--GTAATGATGACGAGGAAGATAATGA------CGAAGAAGCAAGAGGGG---------------AG---------------------------------------- | |
| droEle1 | scf7180000491217:392919-393131 + | GGAC-------------------GATGAGGATGTAG------------CTGAAGACAACGA---GGA--TGGAACTAGGGAG---GAGAAAAACA------ACGAAGACGAAGAC------------------GAGGAC------------------------GACGAAG---------------------------ACGAAGAC---G------AT---------------------AATGACGAAGACAAGGACGAT------------------------------GA---------------TG------------ACGA-GGACGAAGAAGATACTGATAAGGACGATGAAAAGG------------------------------------------------------------------------------------A---------------GG---ATGAGGATGA----CAAAGAAGA------------TGGGGAGAACGACGA--------------CGAGGATACAAATGG---------------------------------------GGA | |
| droRho1 | scf7180000766551:36471-36677 + | GGAT---GCA-------------ACTGGGGAAGAAAATGGTGACGAAGACGAGAACGAA-----------------------------GAAGACG------AAAACGACGATGAC------------GAGAACGACG---------------------------------------------------ATGACGAGAACGACGAT---GAC------GAGA------------------------------------------------ACGACGATGACGAGAACGAC---GAAGAC---------GAG---A-------------A------TGGCAAAGACGACGA---------------------------------------------------------------------TGAC------GAAGACAA---------------TACTGACAAGG---ACGATGAA--GATG--AGAATG---------------AGGA------TGAGGATGACGAAGA--------------------------------------------------------AGA | |
| droBia1 | scf7180000301505:149467-149662 + | AGAT-------------------AACGATGAGG---------------AGGATGATAATAA---GAAGA--------ACGAT------ACTGGTG------GTAAAGATGAAGAT------------------AATG---------------------ACGACGACG---------------------------ACGATGACGAT---G------ATG---------------------ATG---------------------------ATGATGATGATGACGA---------------------CG------------ACG----------------------ACGACGACGACGA---------------------------------------CGACGACGA---CGACGA---TGAT---------GAT------------GATGACGA---CGACG---ACGA----------CGAAGATG---------------AAGA------TGACGAAGACGAAGATG---------------AA-------------------------------------GAC | |
| droTak1 | scf7180000410795:34-219 - | CGAC-------------------GACGACGACG---------------ACGACGACGAG-------A--C---------GAC---GACGACGACG------ACGACGACGACGAC------------------GACG---------------------------ACG---------------------------ACGACGACGAC---G------ACG---------------------ACG---------------------------ACGACGACGACGACGACGACGA---CGAC---------GA----------------CGA------CGA------------------------------------------------------------CGACGACGA---CGACGA---CGAC---------GAC------------GACGACGA---CGACG---ACGA----------CGACGACGA------------CGACGA------CGACGACGACGACGACG---------------AC---------------------------------------- | |
| droEug1 | scf7180000409466:1012637-1012845 + | A--T-------------------AGCGTCGAAG---------------ATGAAGATGAAGC---GGA--T---------GCC---GCCGACGATG------CC---------------AGCAATGCGGCTGA---------------------------------------------------GGAGGAGCATGCAGATGATGAG---GAT------ACGGAT---------------GGCAA---------------------------------------CAATGAG---GAGGAGGACGAG----------------------GACGA------------------------------------------------------------------------------CGATGACCAGGAGGAGGAGGACGACGA------CGAGGATGACTCCGACAGCA---CTGCGGAA--CAGG--AAAAAGAGC------------CCGA------GGCCGAGACCGAAGATG---------------AG---------------------------------------- | |
| dm3 | chr2R:2618293-2618500 - | GGAT-------------------GACGAAGAAG---------------ATGATAATAACGG---GAA--------------------------------------------------------------------------------------------------CG---------------------------------------------------------------------------------ACGAGGACGAC------------------------------GA---------------------------GGACG-AAGAAGAAGATGACAATAATGATGACGATGACGAGGATGATGAAGATGATAATAATGGAGACGACGAGGATGACGAAGAGGACAATAATGAAAACGAAGATGAAGATAATGA------------------------TG---GGGACGAC--GATG--ACGGAGAT------------GACAA------TGAGGACGACAAGGACG---------------AG---------------------------------------- | |
| droSim2 | 2r:3483292-3483494 - | A-AT-------------------GATGATGAAA---------------ATGGGGACGACGG---GGA--T---------GAC---GAAGATGATA------ATAATGAGGACGAC------------------GAGGATGACGAAGAAGATGATAATAACGGGAACG---------------------------------------------------------------------------------ACGAGGACGAT------------------------------GA---------------------------TGACG-AAGATAAAGATGACAATAATGATGACGACGACGAGGATGATGAAGACGA---------------------------------------------------------------------------------------TA---ATAAAATG--GACG--ACGAGGATA------------ACGA------AGAGGA-----------------CAATAATGA---------------------------------------GGA | |
| droSec2 | scaffold_1:291045-291253 - | GGAT-------------------GATGATGAAA---------------ATGGGGACAACGG---GGA--T---------GAC---GAAGATGATA------ATAATGAGGATGAC------------------GAGGATGACGAAGGAGATGATAATAACGGGAAAG---------------------------------------------------------------------------------ACGAGGACGAG------------------------------GA---------------------------TGACG-AAGATGAAGATGACAATAATGATGACGACGACGAGGATGATGAAGACGATAATAAAAGAGG---------------------------------------------------------------------------------CGACGAG--GATG--ACGAAGAGA------------ACAA------TAATGAGGACAAAG------------------------------------------------------------ | |
| droYak3 | 2L:15355119-15355311 - | GGAT-------------------GACGAAGAAG---------------AAGATGATAATGG---GAA--C---------GAC---GAAGACGATA------ATGAAGATGACAAT------------------AACGAC------------------------GACG---------------------------AGAATGACGAA---G------AGG---------------------AAG---------------------------ATGAAGAAGAAGATAATGAT---GACGAC---------GAG---G-------------A------TGA------------------------------------------------------------CGGGGAAGA---TGACAA---TGAG---------GAC------------GACAAGGA---CGAAG---ATGA----------TGAAGAAGA------------TAATAA------TGATGACGACAAGGACG---------------AG---------------------------------------- | |
| droEre2 | scaffold_4929:19927199-19927386 + | G-AT-------------------GACGAGA------------------ATGGGGACGACGA---CGA--T---------GAC---GAGAATGACGAAGAGAATGGAGACGACGAC------------------GAGGAT------------------------GACGAAGA------------GAA---------CAATG---------------------------------------------------------AAGAAGACGAAGATGAAGAAGAAGATAATGAT---GACGAC---------GAG---G-------------A------------------------------------------------------------------------------------------TGAC------GGAGATGGTGACAATGA---------------GG---AAGAGGATGA----CGAAGAAG---------------ATAA------TAATGA-----------------TGAGGATGA---------------------------------------CGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:33 PM