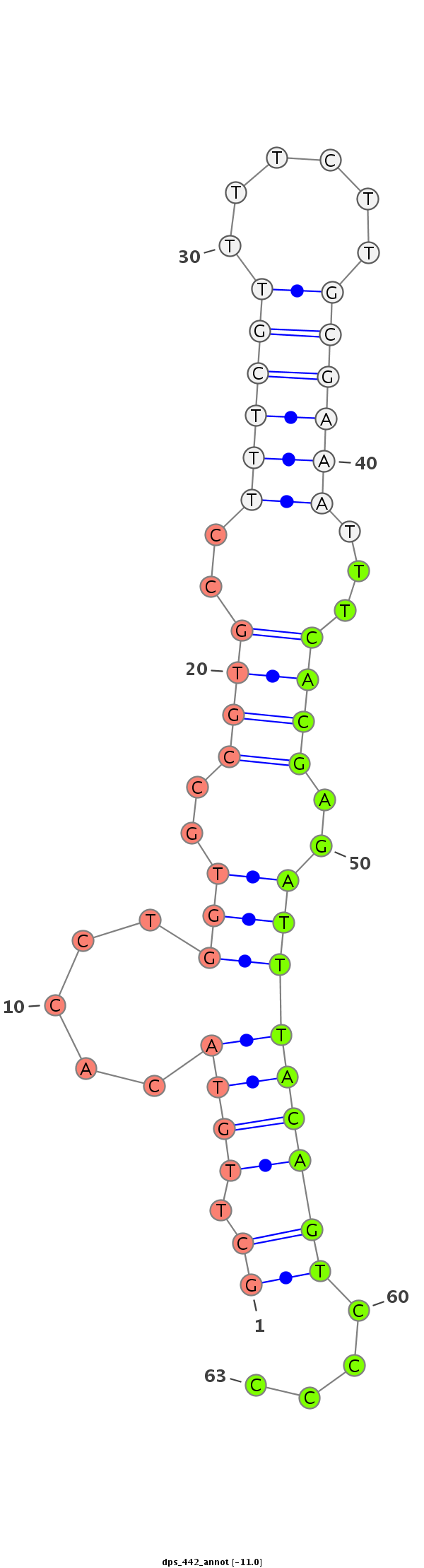
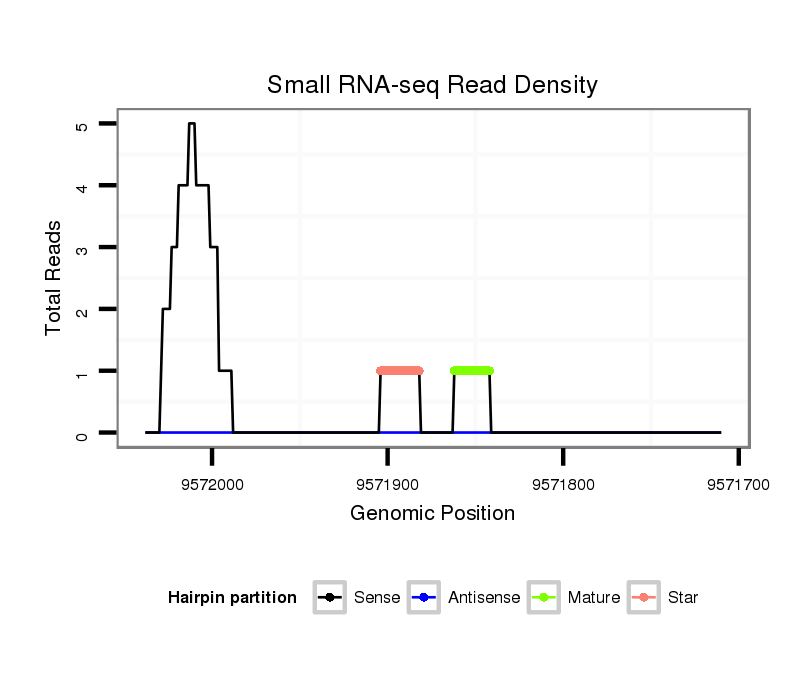
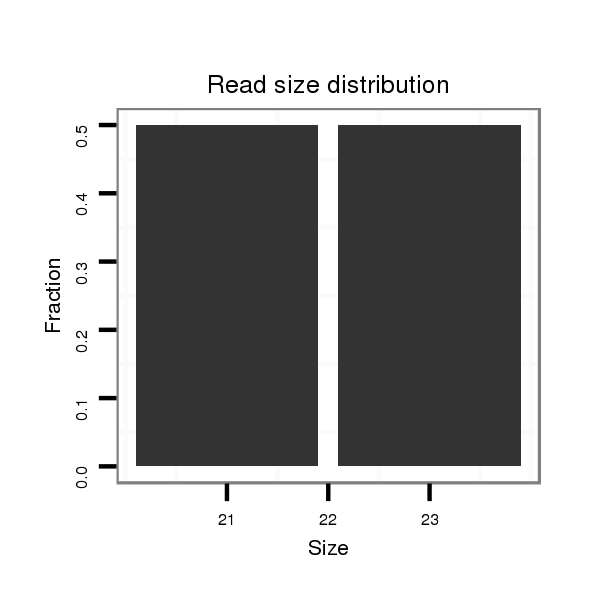
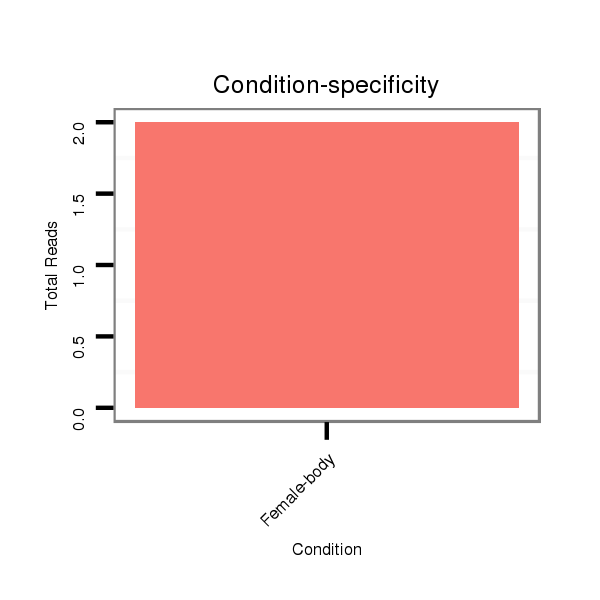
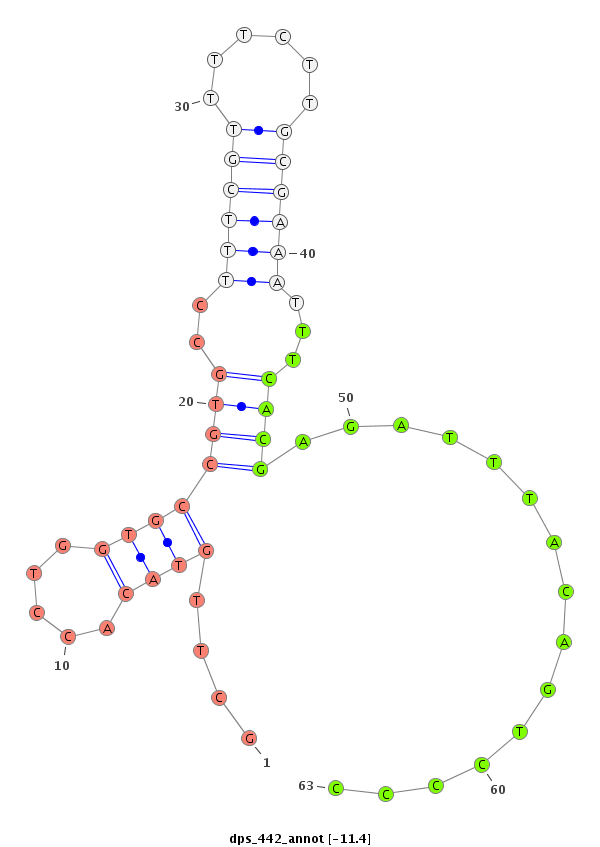
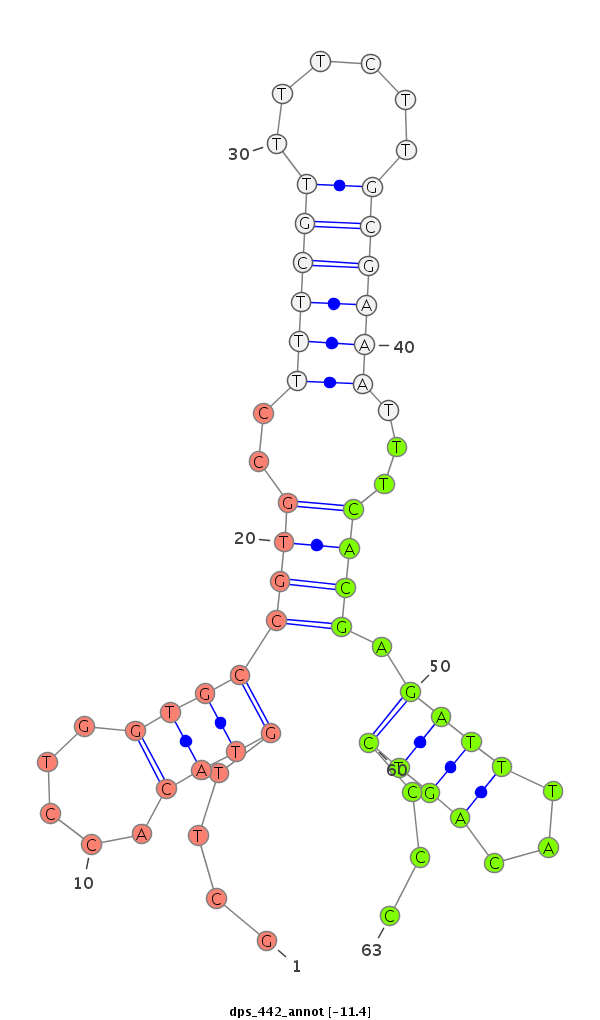
ID:dps_442 |
Coordinate:2:9571760-9571988 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
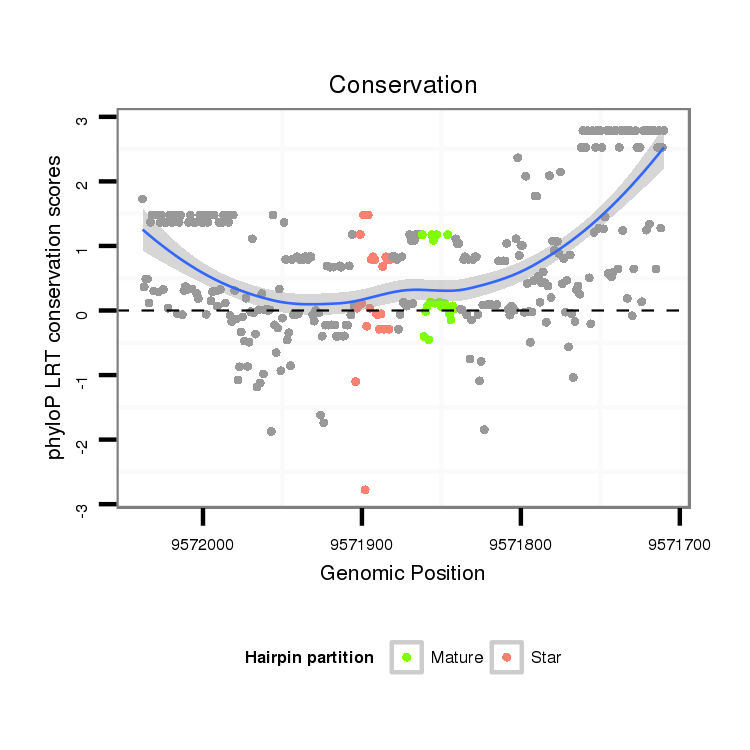
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -11.4 | -11.4 | -11.2 |
 |
 |
 |
exon [Dpse\GA19837:8]; CDS [Dpse\GA19837-cds]; CDS [Dpse\GA19837-cds]; exon [Dpse\GA19837:10]; intron [Dpse\GA19837-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGCGTCACTATCAGCGGAGCATCGAGAAGCATGGCGAGGACGAGAAGAGGGTAAGTGTTAGAGTGTGTGTGTGTGTGTGTTATTTGTGCCGATACATACCCCTTGATTATTGGCAGTCGTCGCGTCGAGCAGATGCTTGTACACCTGGTGCCGTGCCTTTCGTTTTCTTGCGAAATTTCACGAGATTTACAGTCCCCTGCCGCATGGGCTTAGACCCAAAGGGTTTAATCTAACCTCTAAGCTGCTATAATTATATTTTCTACCCGATCTCGAACACAGCTCATGCACTGCATCACAAAGCTGTTCAATTTGCCCATCAAATTCGAGCA **************************************************************************************************************************************((.((((.....(((..((((..((((((......))))))...))))..)))))))))....************************************************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
GSM343916 embryo |
SRR902010 ovaries |
SRR902011 testis |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................GCGTCGAGCAGATGCTTGTCC........................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................TTCACGAGATTTACAGTCCCC.................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............GGAGCATCGAGAAGCATGGCGAGGACG............................................................................................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................ATGGCGAGGACGAGTAGAGGCT..................................................................................................................................................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................GCTTGTACACCTGGTGCCGTGCC............................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................GAAGCATGGCGAGGACGAGAAGAGG....................................................................................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........ATCAGCGGAGCATCGAGAAG............................................................................................................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................CATCGAGAAGCATGGCGAGGACG............................................................................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................................................................................AGCACAAAGCTGTTCAATT................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........TCAGCGGAGCATCGAGAAGCATGGCGA.................................................................................................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................GCGAGGACGAGAAGAGGCT..................................................................................................................................................................................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GGGTAATTGTAAGAATGTGTGT................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................GCATGGCGAGGACGAGAAGAGGCTC.................................................................................................................................................................................................................................................................................... | 25 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................CATGGCGAGGACGAGAAGAGGC...................................................................................................................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................GATTGGCGGTCGTCGCGTC........................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................AACCTGTTGAATTTGCCCTTCA.......... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................TGGTAAGTGTAAGAGTGTGTCT................................................................................................................................................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................TGTTTGTGTGTTGTTTGTGCC............................................................................................................................................................................................................................................... | 21 | 2 | 11 | 0.27 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TGGTTCACCTGGTGCCATGC............................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CGCGTTGACCAGTTGCTTGT............................................................................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................GCTGCTTTTACACGTGGTGC.................................................................................................................................................................................. | 20 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................AAGGCGAGGGCGAGAGGAGGG...................................................................................................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
ACGCAGTGATAGTCGCCTCGTAGCTCTTCGTACCGCTCCTGCTCTTCTCCCATTCACAATCTCACACACACACACACACAATAAACACGGCTATGTATGGGGAACTAATAACCGTCAGCAGCGCAGCTCGTCTACGAACATGTGGACCACGGCACGGAAAGCAAAAGAACGCTTTAAAGTGCTCTAAATGTCAGGGGACGGCGTACCCGAATCTGGGTTTCCCAAATTAGATTGGAGATTCGACGATATTAATATAAAAGATGGGCTAGAGCTTGTGTCGAGTACGTGACGTAGTGTTTCGACAAGTTAAACGGGTAGTTTAAGCTCGT
************************************************************************************************************************************((.((((.....(((..((((..((((((......))))))...))))..)))))))))....************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
SRR902011 testis |
M022 male body |
GSM444067 head |
SRR902010 ovaries |
SRR902012 CNS imaginal disc |
M062 head |
GSM343916 embryo |
M059 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................GCGTACGGGACGTAGTGTTG.............................. | 20 | 3 | 12 | 232.92 | 2795 | 891 | 759 | 1069 | 9 | 55 | 5 | 6 | 0 | 0 | 1 |
| .......................................................................................................................................................................................................................................................................................GCGTACGGGACGTAGTGTT............................... | 19 | 2 | 3 | 135.00 | 405 | 325 | 24 | 41 | 2 | 10 | 1 | 1 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TGGCGTACGGGACGTAGTGT................................ | 20 | 3 | 8 | 103.13 | 825 | 639 | 135 | 16 | 0 | 26 | 5 | 0 | 4 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TGGCGTACGGGACGTAGTGTT............................... | 21 | 3 | 3 | 87.67 | 263 | 203 | 41 | 9 | 0 | 3 | 6 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TTGCGTACGGGACGTAGTGTT............................... | 21 | 3 | 2 | 64.50 | 129 | 87 | 34 | 0 | 0 | 7 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................GTACGGGACGTAGTGTTG.............................. | 18 | 2 | 5 | 50.00 | 250 | 110 | 4 | 1 | 124 | 10 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................................................................TTGCGTACGGGACGTAGTGT................................ | 20 | 3 | 11 | 17.36 | 191 | 130 | 55 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................GCGTACGGGACGTAGTGT................................ | 18 | 2 | 10 | 9.20 | 92 | 84 | 1 | 3 | 0 | 3 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................AGTACGGGACGTAGTGTTG.............................. | 19 | 2 | 1 | 9.00 | 9 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................TGTACGGGACGTAGTGTT............................... | 18 | 2 | 4 | 8.50 | 34 | 32 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................CGTACGGGACGTAGTGTT............................... | 18 | 2 | 12 | 8.33 | 100 | 93 | 0 | 0 | 5 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CGAGTATGTGACGTAGAGT................................ | 19 | 2 | 1 | 4.00 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................TGTACGGGACGTAGTGTTG.............................. | 19 | 3 | 20 | 3.25 | 65 | 35 | 21 | 6 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................GCGTACGGGACGTAGTGTTT.............................. | 20 | 2 | 1 | 3.00 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................AAGTACGGGACGTAGTGTT............................... | 19 | 2 | 1 | 3.00 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................GGCGTACGGGACGTAGTGTTT.............................. | 21 | 3 | 3 | 2.33 | 7 | 4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CGAGTATGTGACGTAGAGTG............................... | 20 | 3 | 4 | 2.25 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................TTGTACGGGACGTAGTGTT............................... | 19 | 3 | 20 | 1.05 | 21 | 20 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................CGTACGGGACGTAGTGTTT.............................. | 19 | 2 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CGCGTACGGGACGTAGTGTT............................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................AGTACGGGACGTAGTGTT............................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TCGCGTACGGGACGTAGTGT................................ | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CGAGTATGTGACGTAGAG................................. | 18 | 2 | 3 | 1.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................GCGTACGGGACGTAGTGTTGC............................. | 21 | 3 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CGAGTATGTGACGTAGAGTGT.............................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................TGCGTACGGGACGTAGTGTTT.............................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CTAAATGTCAGGCGGCG................................................................................................................................. | 17 | 2 | 8 | 0.50 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................TGGAGATTCGACGACAATA.............................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................................................................GCGTACGGGACGTAGTGTTGCG............................ | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................AAGTACGGGACGTAGTGTTG.............................. | 20 | 3 | 6 | 0.33 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................CCTAGATGTCAGGGGACG................................................................................................................................. | 18 | 2 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CGCGTACGGGACGTAGTGTTG.............................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................GGAGTACGGGACGTAGTGTTG.............................. | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AGCGTACGGGACGTAGTGTT............................... | 20 | 3 | 13 | 0.31 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TAGCGTACGGGACGTAGTGTT............................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................CCTAAATGTCAGGTGACG................................................................................................................................. | 18 | 2 | 8 | 0.25 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CTAGATGTCAGGGGGCG................................................................................................................................. | 17 | 2 | 14 | 0.21 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TCTAAATGTCAGAGCACG................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................ATGTACGGGACGTAGTGTT............................... | 19 | 3 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................TAGTACGGGACGTAGTGT................................ | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................GTACGGGACGTAGTGT................................ | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CCAGTACGGGACGTAGTGT................................ | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................GTGTACGGGACGTAGTGTTG.............................. | 20 | 3 | 10 | 0.20 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................ATTACGGGACGTAGTGTT............................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................ATTGGAGATTCGTCGAGA................................................................................. | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................GCGTACGAGACGTAGTGTTG.............................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CTAAATGTCAGGCGGC.................................................................................................................................. | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................ACGGGACGTAGTGTT............................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................CGTACGAGACGTAGTGTTG.............................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CTATATGTCAGGTGACG................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................ATTACGGGACGTAGTGTTG.............................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CTAAATGTCAGGTGTCG................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:9571710-9572038 - | dps_442 | TGCGTCACTATCAGCGGAGCATCGAGAAGCATGGCGAGGACGAGAAGAGGGTAAGTG-TTA-------------------GAGTGTGTGTGTGT------GTGTGTTATTTGTGCCG-ATACATACCCCT--TGATTA-----------------------------------------------------------TTGGCAGTCGTCGCGTCGAGCAGATGCTTGTACACCTGGTGC--CGTGCCTTTCGTT----------TTCTTGCGAAATTTCAC----------------------GAGATTTACAGTC-----------------------------------------------------------------------------------------------------------------CCCTGCCGCATGGGCTTAGACCCAAA---GGGTTTAATCTAACCT-CTAAGCTGCTATAAT-----TATA-T---TTTCTACC-CG----------ATCTCGA-------------ACACAGCTCATGCACTGCATCACAAAGCTGTTCAATTTGCCCATCAAATTCGAGCA |
| droPer2 | scaffold_0:10908047-10908384 + | dpe_470 | TGCGTCACTATCAGCGGAGCATCGAGAAGCATGGCGAGGACGAGAAGAGGGTAAGTG-TTA-------------------GAGTGTGTGTGTGTGTGTGTGTGTGTTGTTTGTGCCG-ATACATACCCCT--TGATTA-----------------------------------------------------------TTGTCTGTCGTCGCGTCGAGCAGATGCTTGTGCACCTGGTGC--CGTGCCTTTCGTT----------TTCTTGCGAAATTTCAA----------------------GAGATTTACAGTC-----------------------------------------------------------------------------------------------------------------CCCTGCCGCATGGGCTTAGACTCAAAGCTGGGTTTAATCTAACCT-CTAAGCTGCTATAAT-----TCTA----TTTTCTACC-CG----------ATTTCGA-------------ACACAGCTCATGCACTGCATCACAAAGCTGTTCAATTTGCCCATTAAATTCGAGCA |
| droWil2 | scf2_1100000004943:8688823-8689185 + | dwi_3371 | TGCGTCACTACCAACGGAGCATTGAACGACATGGACAGGATGAGAAAAGAGTAAGTT-TTG-TCGCCGTCTCGATTTCTATAACTTGTGTAATT------ACTTGCTCTATATGAC-----------------------------------------------AATGC---------------------CCCTATTCCAAAAAATTGCCAATTCTCATAGATCATTGTTTAA---------------------------------------------------------------------------------------------------------------ATGACCTTTCACTCTCTAAAGACAGACAGAGAGAGAGAGAAAGAGAGTGAGCACTTACAACTATGCAAACACCTGTCATTACACCTCTCTGCCT------------------------------CTCTCATACTAATCTGGGTTACCCTTTTTATT----TTTTCTACC-TGTGTGTGTGTGTTTTT----------------TCCAGCTTCTTCACTGTATCAGCAAACTGTTTAATTTGCCCATCAAATTTGAGCA |
| droVir3 | scaffold_13047:13382118-13382213 + | TCTA------------------------------------------------------------------------------------------ATCCATAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGCTGT--------------T-----TTTTCGACTACTACCTG----TC-----------------CACGCAGCTGTTGCACTGCATCACAAAGCTGTTTAACCTGCCCATCACATTCGAGCA | |
| droMoj3 | scaffold_6540:10617455-10617509 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-----------------------------------------------------------------------GCAGCTGTTGCACTGCATCACAAAGCTGTTTAACCTGCCCATCACATTCGAGCA | |
| droGri2 | scaffold_14906:4307224-4307278 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-----------------------------------------------------------------------GCAGCTGCTGCACTGCATCACAAAGCTGTTCAACCTGCCCATCACATTCGAGCA | |
| droAna3 | scaffold_13340:8313710-8314005 - | TGCGCCACTACCAGCGCAGCATCGAAAAGCATGGCGAGGACGAAAAACGGGTGAGTG-TATTTCACCCTTGCCCTGTCC-------TTTTACCC------TTTCTCCCTGTCTGCC-----------------------------------------------AATCC---------------------CCCAACTTCAAA--------------------------TTAA------------------------GCTCAAGGCATTATGCGAAATTCCCC----------------------AAGGTCACCAGTGTGTATCT-------------------------------------------------------------------------------------------CATACCGTTGCACCTGCTTGCCG-------------------------TTAATCACATCTCCTAATCTCGG---------------------TCTACC-TG----------TTTCTGT--TTCACC-ATCCACACAGCTGTTGCACTGCGTTACCAAACTGTTCAATCTGCCGATCAAATTTGAGCA | |
| droBip1 | scf7180000396708:523410-523712 + | TGCGCCATTACCAGCGCAGCATCGAAAAGCATGGCGAGGACGAAAAACGGGTGAGTG-TAATTCACCCTCACCACGTCCACCCCTTCGTTACTC------CTGCGCCCTGTCTACC-----------------------------------------------AAGTC---------------------CCCCAATTCAAA--------------------------TGAA------------------------ACTTAAGCCATTTTGCGAAATTCCCC----------------------AAGGTCACCAGTGTGTATCT-------------------------------------------------------------------------------------------CATGCCGTTGCACCTGCCTGCAG-------------------------TTAATCACATCTCCTAATCTCGG---------------------TCTACC-TG----------TTTTCGT--TTTACC-ATTCACACAGCTGTTGCACTGCGTTACCAAACTGTTCAATCTGCCGATCAAATTTGAGCA | |
| droKik1 | scf7180000302307:491282-491336 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-----------------------------------------------------------------------ACAGCTGGTGCATTGCATGAACAAGCTGTTCAACCTGCCCATCAAATTCGAGCA | |
| droFic1 | scf7180000453906:777648-777746 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCAAGCTGGCTTAAT-----CACC-CCTTTTTCTACC-TG----------CTCTCCCA-------C---CAAACAGCTGCTGCACTGCATCACCAAGCTGTTTAATCTGCCAATCAAATTCGAGCA | |
| droEle1 | scf7180000491280:191201-191302 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAAGCTGGC-TAAT-----CACCTTCGTTATCTACC-TG----------CTCTCCCC----AAAA---CAAACAGCTGCTGCACTGCATCACCAAGCTATTCAATCTGCCCATCAAATTCGAGCA | |
| droRho1 | scf7180000780099:26802-26900 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAAGCTGGCTTAAT-----CACC-TTGTTTTCTACC-TG----------CTCTCCAA-------A---CAAACAGCTGCTGCACTGCATCACCAAGCTATTCAATCTGCCCATCAAATTCGAGCA | |
| droBia1 | scf7180000302402:4603755-4603857 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAAGCCATCTTAAT-----CACC-TCTTTCTCTACC-TG----------CTCTCCCC----AAA-AC-CCAACAGCTGCTGCACTGCATCACCAAGCTGTTCAACCTGCCCATCAAATTCGAGCA | |
| droTak1 | scf7180000414118:10989-11074 + | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTCTACC-TG----------CTCTCGCAATCAAAA-ACCAAAACAGCTGCTGCACTGCATCACCAAGCTGTTCAACCTGCCCATCAAATTCGAGCA | |
| droEug1 | scf7180000409473:272735-272833 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAAGCTGTCTTAAT-----CACA-TTGTTCTCTACC-TG----------CTTTCGCA-------C---AAAACAGCTGCTGCACTGCATCACCAAGCTGTTCAATCTGCCGATCAAATTCGAACA | |
| dm3 | chr3R:19077630-19078020 + | TGCGCCACTACCAGCGCAGCATTGAGAAGCACGGCGAGGACGAACAACGGGTGAGCTATTA-------------------CCCTC----------------------ATTGGCACCGCCATCGCATCCCTTTTGATCCACTGCTTACTGCCTTCCTATCATTCAAGTCCCCCATTTTCCTAACCATTTA--AT--------------------------CACCCCTATGTACCTGACCTGG-CTGGCTTTCCCTTACTTA----ACCTTGTGAAATTCGCAAACATAT----GTACCTCCGCCAAGGTCACTTTTCTCGACTTTAAGTCGCTCTTCTTTTCCGCTGACCTTTCACT----CA---------------------------------------------------------------------------CACGGTCTTTTG--------------------------CTAATCTTTCTTAAT-----CACC-TCTTTCGCTACC-TG----------CTCTCCCG-------T---CGAGCAGCTGCTGCACTGCATCACCAAGCTGTTCAACCTGCCGATCAAATTCGAGCA | |
| droSim2 | 3r:18620245-18620343 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAATCTTTCTTAAT-----CACC-TCTTTCGCTACC-TG----------AACTCCCG-------C---CGAGCAGCTGCTGCACTGCATCACCAAGCTGTTCAACCTGCCGATCAAATTCGAGCA | |
| droSec2 | scaffold_0:19442511-19442609 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAATCTTTCTTAAT-----CACC-TCTTTCGCTACC-TG----------AACTCCCG-------C---CGAGCAGCTGCTGCACTGCATCACCAAGCTGTTCAACCTGCCGATCAAATTCGAGCA | |
| droYak3 | 3R:19952436-19952836 + | TGCGCCACTACCAGCGCAGCATTGAGAAGCACGGCGAGGACGAACAACGGGTGAGTTATTA-------------------CTCTCC---------------------GTTTGCAACGCATTCGCATCCCT-TTGATCCCCTGCTTACGGTCGTCTATTTCTTTAATTCCCTCATTTTCGTAGCCATGTACCCT--------------------------CACTCCTATGTACCTGACCTGACCTGGCTTTCCCTTACTTA----ACCTTGTGAAATTTGCATACATATGTATGTACCTCCGCCAAGGTCACTTTTCGCGACTTTAAGTCGCTCTTCTT-TCCGCTGACCTTTCACTCACTCA---------------------------------------------------------------------------CACGTTCTTTTG--------------------------CTAATCGTTCTTAAA-----CACC-TCTTTCGCTACC-TG----------CTCTAACG-------C---CGAGCAGCTGCTGCACTGCATCACCAAGCTGTTCAACCTGCCGATCAAATTCGAGCA | |
| droEre2 | scaffold_4820:8976446-8976831 - | TGCGCCACTACCAGCGCAGCATTGAGAAGCACGGCGAGGATGAACAACGGGTGAGCTATTA-------------------TTCTC----------------------GTTCGCCCCGCATACGCATCCCT--TGATCCCCTGCTTACAATGGTCTTTTCATTGAATTCCCTCATTTTTCTACCCAT-----AT--------------------------CACCCCTATATACCTGACCTGG-CTGGCTTTCCCTTACTTA----ACCTTGTGAAATTTGCATACAT--------ACATCCGCCAAGGTCACTTTTCGCGACTTTAAGTCGCTCTTCTTTTCCGCTGACCTTTCACTCGCCCA---------------------------------------------------------------------------CACGTTCTTTGA--------------------------CTAATCGTACTTAAT-----CACC-TCTCTCGCTACC-TG----------CTATCCCG-------C---GGAGCAGCTGCTGCACTGCATCACCAAGCTGTTCAACCTGCCGATCAAATTCGAGCA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:23 PM