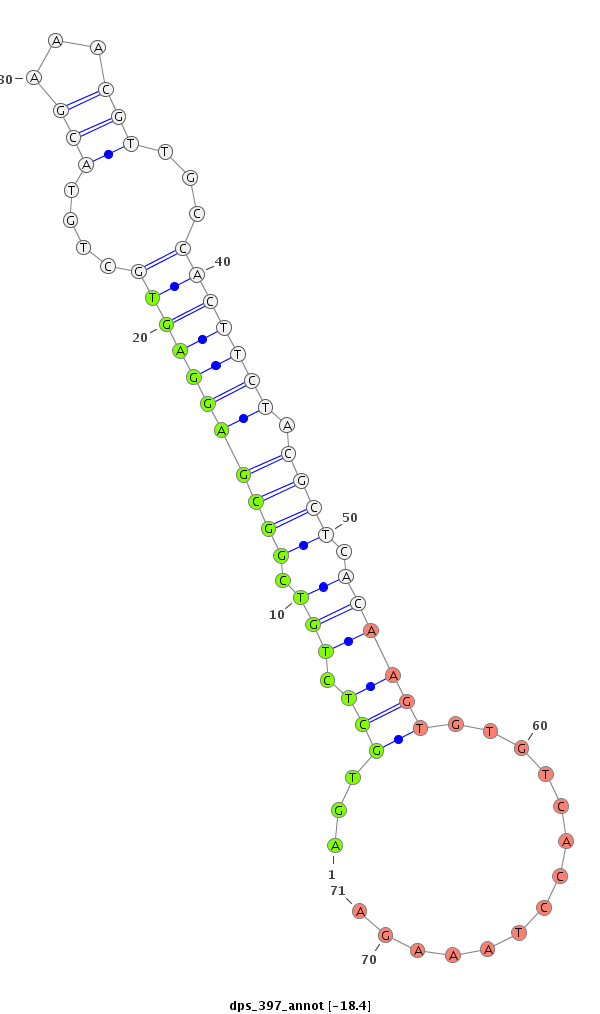
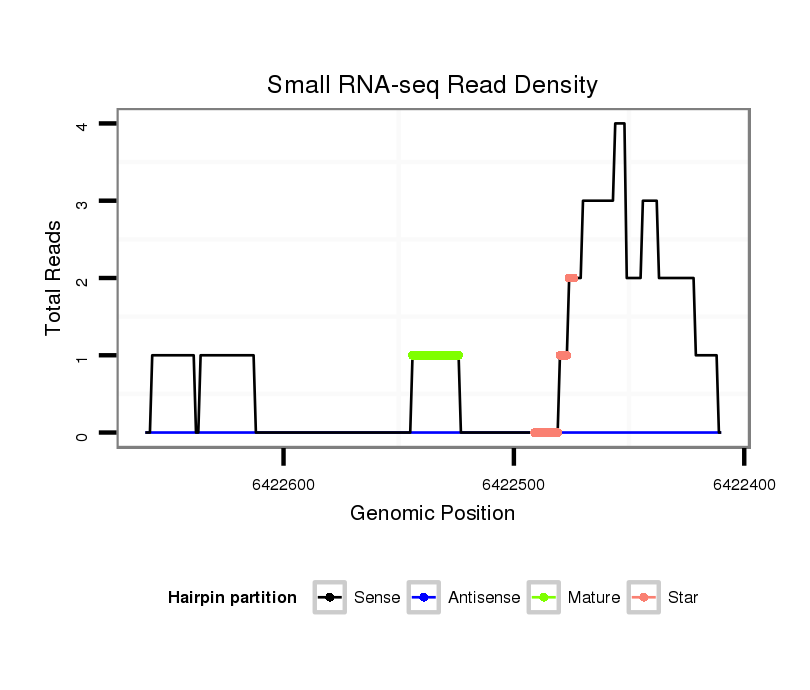
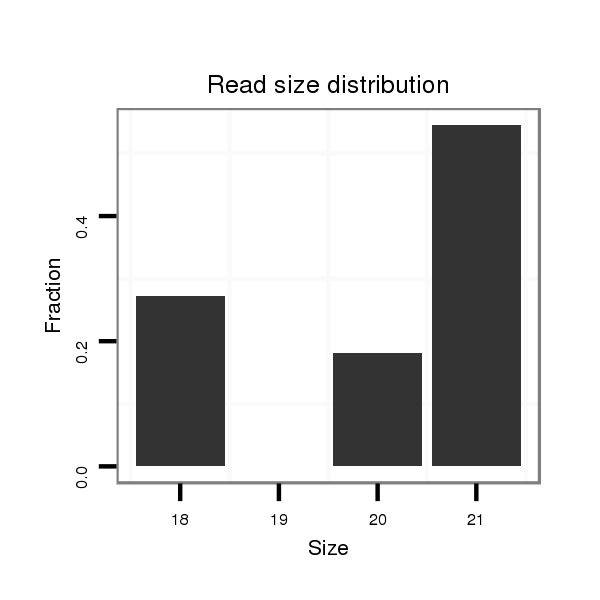
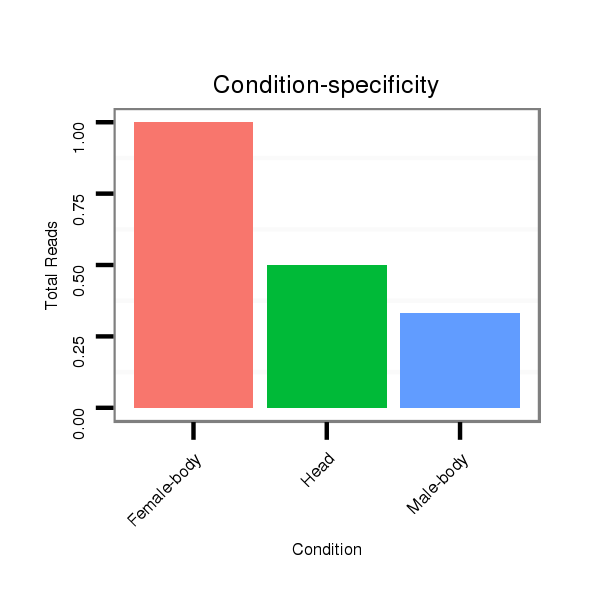
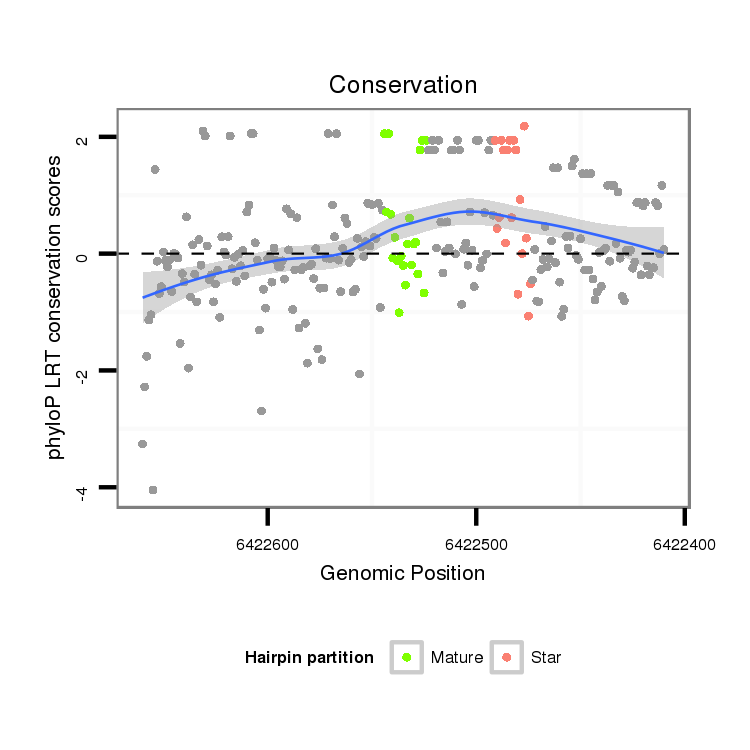
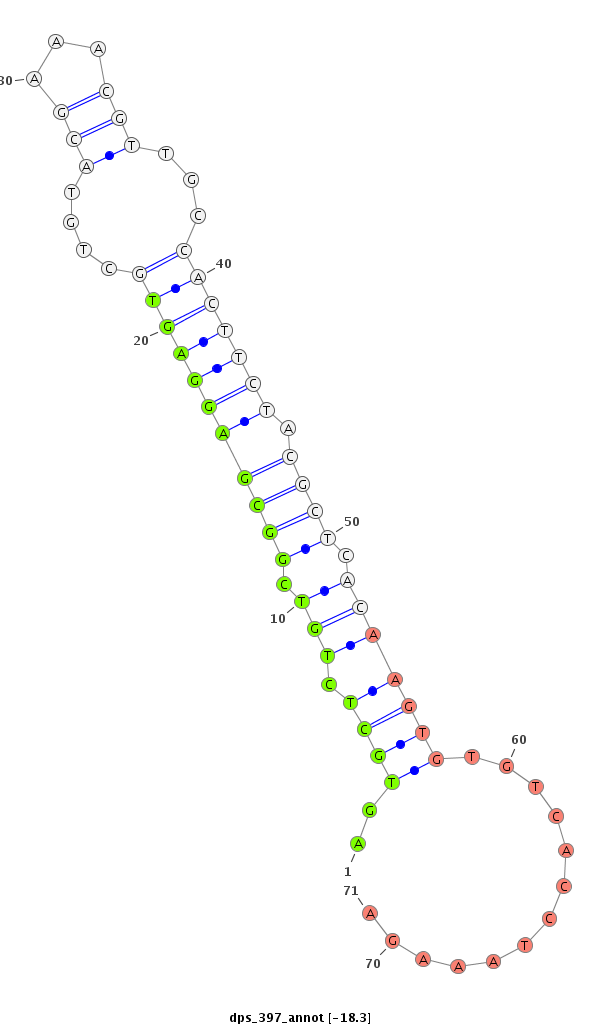
ID:dps_397 |
Coordinate:2:6422460-6422610 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -18.3 | -18.1 | -18.1 |
 |
 |
 |
exon [dpse_GLEANR_4299:1]; CDS [Dpse\GA26723-cds]; intron [Dpse\GA26723-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGACACCACAGACGGAGCAGGGCAGGCAGTGCCAGCCGGTCGGTGGACAGGTAAGAGTCAGTGTGGATCTCACCTTGAGCCCATTGTTAACTTTCTCTCGTTACGCACTTTTCTCCAGTGCTCTGTCGGCGAGGAGTGCTGTACGAAACGTTGCCACTTCTACGCTCACAAGTGTGTCACCTAAAGAGATGTCAGACAAAACGAGTAATAATAATAGCTCGGATTATTACTCATTCGATACATACATGTAC ********************************************************************************************************************...(((.(((.(((((((((((....(((...)))...))))))).)))).))))))..............**************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
GSM343916 embryo |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................TTGGCGAGGAGTGCGGTTC........................................................................................................... | 19 | 3 | 17 | 5.76 | 98 | 52 | 45 | 0 | 1 |
| ............................................................................................................................GTTGGCGAGGAGTGCGGTTC........................................................................................................... | 20 | 3 | 6 | 4.67 | 28 | 7 | 21 | 0 | 0 |
| ............................................................................................................................................................................................................GTAATAATAATAGCTCGGATTATT....................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................GGACAGTGCTCTGTCGGCGAGGAGTGCT............................................................................................................... | 28 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................GTCGGCGAGGAGTGCGGTTC........................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................AGAGATGTCAGACAAAACGAGTAAT.......................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................TGGACAGTGCTCTGTCGGCGAGGA.................................................................................................................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................GGCAGTGCCAGCCGGTCGGTGGAC........................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GCTCGGATTATTACTCATTCGAT............ | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...CACCACAGACGGAGCAGGG..................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................ACTCATTCGATACATACATGT.. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................AGTGCTCTGTCGGCGAGGAGT.................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................GTCAGACAAAACGAGTAAT.......................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................CTAAAGAGATGTCAGACAAAACGA............................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GTAATAATAATAGCTCGGA............................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................GTTGGCGAGGAGTGCGGT............................................................................................................. | 18 | 2 | 6 | 0.67 | 4 | 1 | 3 | 0 | 0 |
| ...............................................................................................................................GGCGAGGAGTGCTGTTCT.......................................................................................................... | 18 | 2 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 |
| .................................AGCCGGACGGTGGCCATGTAA..................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................AAGTGTGTCACCTAAAGA................................................................ | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 |
| .............................................................TGTGGATCTCACCTTGAGCC.......................................................................................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................GTCGGCGAGGAGTGCGGTT............................................................................................................ | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................TTGGCGAGGAGTGCGGTTCG.......................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................GTTGGCGAGGAGTGCGGTT............................................................................................................ | 19 | 3 | 20 | 0.30 | 6 | 2 | 4 | 0 | 0 |
| .....................................GGGCGGGGGACCGGTAAGAG.................................................................................................................................................................................................. | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 |
| ...................................................................................................................................................................GCGCTCAAGTGTGTTACCT..................................................................... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................GTGGGCGAGGAGTGCGGTTC........................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................CCGTTGGCGAGGAGTGCGG.............................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
TCTGTGGTGTCTGCCTCGTCCCGTCCGTCACGGTCGGCCAGCCACCTGTCCATTCTCAGTCACACCTAGAGTGGAACTCGGGTAACAATTGAAAGAGAGCAATGCGTGAAAAGAGGTCACGAGACAGCCGCTCCTCACGACATGCTTTGCAACGGTGAAGATGCGAGTGTTCACACAGTGGATTTCTCTACAGTCTGTTTTGCTCATTATTATTATCGAGCCTAATAATGAGTAAGCTATGTATGTACATG
****************************************************************...(((.(((.(((((((((((....(((...)))...))))))).)))).))))))..............******************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343916 embryo |
V112 male body |
M040 female body |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................ATGCGAGTGTTCACACAGTGG...................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| .........TCTGCGTCGTCCCGACCGTCACT........................................................................................................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................CGAGAGTGCACACAGTAGA..................................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | 2:6422410-6422660 - | dps_397 | AGACAC------------------------------------------C-ACAGACGGAGC------AGGGCAGGCAGTGCCAGCCGGTCGGTGGACAGGTAAGAGTCAGTGTGGATCTCACCTTGAGCC--------C----------ATTGTTAAC-------TTTCTCT--CGTTAC----------GCACTT-TTCTCCAGTGCTCTGTCGGCGAGGAGTGCTGTACGAAACGTTGCCACTTCTACGCTCACAAGTGTGTCACCTAAAGAGATGTCAGACAAAACGAGTAATAATAATAGCTCGGATTATTACTCATTCGATACATACATGTAC |
| droPer2 | scaffold_0:248640-248891 - | AGACAAC-----------------------------------------C-ACAGACGGAGC------AGGGCAGGCAGTGCCAGCCGGTCGGTGGACAGGTAAGAGTCAGTGTGGATCTCAGCTTGAGCC--------C----------AGTGTTAAC-------TTTCTCT--CGTTAC----------GCACTT-TTCTCCAGTGCTCTGTCGGCGAGGAGTGCTGTACGAAACGTTGCCACTCCTACGCTCACAAGTGTGTCACCTAAAGAGATGTCAGACAAAACGAGTAATAATAATAGCTCGGATTATTACTCATTCGATACATACATGTAC | |
| droWil2 | scf2_1100000004943:10595840-10595888 - | ACAAACGTG------------------------------------------CAAAC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAAAGACGGAAAACAAAAACAATAATAATAATA----------------------------------- | |
| droVir3 | scaffold_12855:8204408-8204453 - | TGGAGC------------------------------------------C-GCAGACGCAGC------CGGGCAGGCAGTGCCATGTGGCCAGTAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14906:7552388-7552589 - | AGATC-------------------------------------------C-AACGAC------------GAGAAGAACATGTAAACTTGTTGGTCATGAAGTAAGCTGCTTG--------TAGCCCGAAGC--------C----------ATTTCTCAC-------TTTCTCA--CAT--TTCTT-TTCGTTTATTA-TTTTACAGTGTTCAGATAGCAGTGAATGCTGCACAAAGCGTTGTGCATTCTATCTTCACAAGTGCGTCACGTAAAGGGAAAACAAGCAAACTTCATAATA----------------------------------------- | |
| droAna3 | scaffold_13340:8552841-8553058 - | GTACGCCAG-----------------------AATGA------------AATTTCTG-GGTCAACGGAGCCCAATAAGTGCCGCAACGTTGGTGAAACGGTAATATAACTTACGTCACTC--------CATTTTGCCTCAGTTACCTCA---TATAAG-------TATCT----CGTATTATAT-TTTCTTTATT--ATTTTCAGTGTTATCGGGGCGAGGAATGCTGTACAATGCGCTGCCACTTTTATATGCACAAATGCGTAACATAAATGAACGT----------------------------------------------------------- | |
| droBip1 | scf7180000396708:284601-284836 + | CCTCGGGTAACACTTTCGACATTGTACGCCAGAATGA------------AATTTCTG-GGTCAACGGTGCCCTATAAGTGCCGCAATGTCGGCGAAACGGTAAGATCACTTACGTCATTC--------CATTTTACCTCAGCCACCTCA---AATAAT-------TAT--------TATTAAATCTTTCTTTGTT--AATTTCAGTGCTACCGGGGTGAGGAATGCTGTACAATGCGCTGTCACTTCTACATGCACAAGTGTGTCACATGAGAATAT------------------------------------------------------------- | |
| droKik1 | scf7180000302388:1516940-1517189 + | CTTCAAAGGAAGTTTATAATATTGTCT---------------------C-GCAGGC------------AGCACCACAATGCCAGAATGTCGGAGGACCGGTAAGGATTTCC----------CTTAAAATA--------T----------AATAAACATGAACTATTTTAACT---CTTTT----------CTTTTTATTTTTCAGTGTAACACCAGCACGGACTGCTGTAATTTGCGCTGTCACTTCTACATGCACAGGTGTGTCACCTGAGCGATTTTAGGGGACAATAAGGAAATGTGACAAACCAGCTTATTACCAAAT-------------TAC | |
| droFic1 | scf7180000454106:704430-704642 - | CATCGAACGATGTTTTTGTAGTCGACT---CGAAGGC------------AGAT-TTCGAAT------CGGCTGCTGAATGCCAGCCAGTCGGTGGACCGGTAAGAATACTT----------CTTAGTAAT--------T----------TTAATGAAC-------TTTCTCTACCAATGT----------GTATAT-TTTCCCAGTGCGATCGCGCTGATGAATGCTGCAATTTGCGATGTTTGACGTACATGCACAGGTGTGTCACTTAAAAAGTTCCCA--------------------------------------------------------- | |
| droEle1 | scf7180000491212:1988394-1988609 - | CATCGGATGATGTTTACGATTTTAAAC---CAAGGGCTGGTTTCGGTCCAATTGAAG-GAT------CAGCAGCAGAATGCGGGAATGTCGGGGATCCGGTAAGAAAACTA----------CATCATCAA--------T----------TTGACCAAC-------TTTCACTTTCGATAT----------ATATTTATATTTTAGTGCTCCCGGGCTGAGGAGTGCTGCAATCTGCGATGCCACACGTACATGCACAGGTGTGTCACCTAA------------------------------------------------------------------- | |
| droRho1 | scf7180000767168:4614-4651 - | ATAAAAACT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTATTATTTATTTTATGCATGTATGCAC | |
| droBia1 | scf7180000302402:6549029-6549260 + | AGACGT-------TTTTTTCGTCGGGC---------------------C-AAAGACAAAAG------CAGCGGCAGAATGCGGGCAAACTGGTGAACCTGTAAGTCAATTT----------ATTTATCT---------A----------ATCACCAAC-------TTTCCCTCTCGACTT----------GTACTTATTTTTCAGTGCTCCCGGGCTGAAGAATGCTGCAACTTGCGATGCCACACTTATATGCATAGGTGTGTCACCTGAAAAATATTCAAGCATACGGA---AAGATGACAGC-CAGATAATT---------------------AC | |
| droTak1 | scf7180000415711:1583974-1584170 + | GGATGA----TGTTTTTGCTATTGATC---------------------C-AAAGACAAATT------ATTCCGCAGAATGCCATCAAATCGGTGAACCAGTAAGTTCATTT----------CTTTAACCG--------T----------TTGAGCAAC-------TTTCAATTTCGTTTT----------CTACTAATTTTCCAGTGCTCTCGGGCAGAAGAATGCTGCAATTTGCGATGCCACACTTATTTGCACAGGTGTGTCACCTAAAAA---------------------------------------------------------------- | |
| droEug1 | scf7180000409490:864795-864979 - | AGTTAG-------------------CG-------TG---------------AAGGA------AAAGGTGACTCCAACCTGTAATCAAACCGGTGGACCTGTAAGATCTTTTCT--------CCTCAATAA--------T----------TTGGCCTAC-------TTTATCTCAGGACTT----------ACATTT-TTTTTCAGTGCTCCCGGGCTGAAGACTGCTGCAATTTCCGATGTCATACTTATATTCACAGGTGTGTCACTTAAAGGGA-------------------------------------------------------------- | |
| droYak3 | 3L:11543393-11543448 + | ACAAAAGCA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAATAATAATAATAATAATAATAATAATTCCTATACACACTTGTAG | |
| droEre2 | scaffold_4784:18438829-18438926 + | GGGCAGGCA------------------------------------------GGTA------------------------GTCGGCCAGTCAGACAGAAGTGAATAATCAATATC-------ATTTGATTA--------C----------ATTGATAAC-------GCTTTTT--CGTGCC----------GCTTTT-TTCTCCATTCCT--------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:20 PM