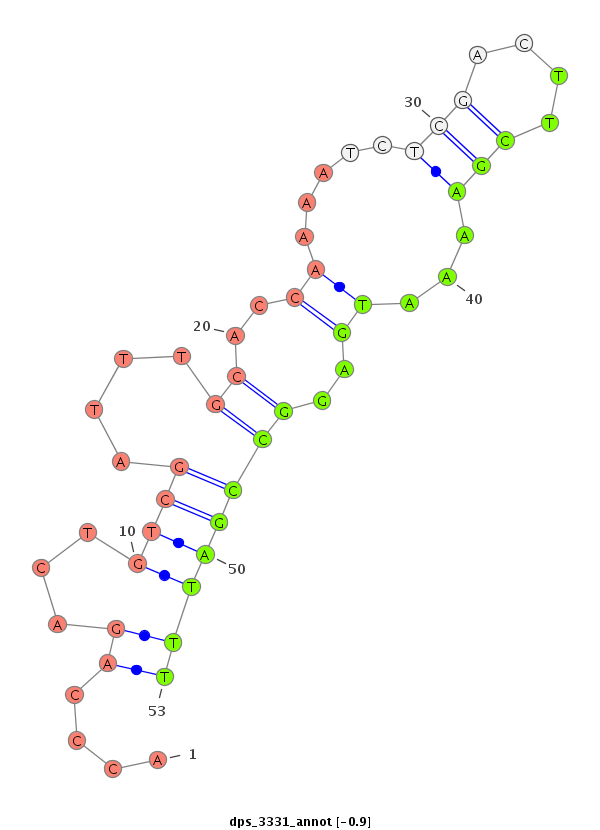
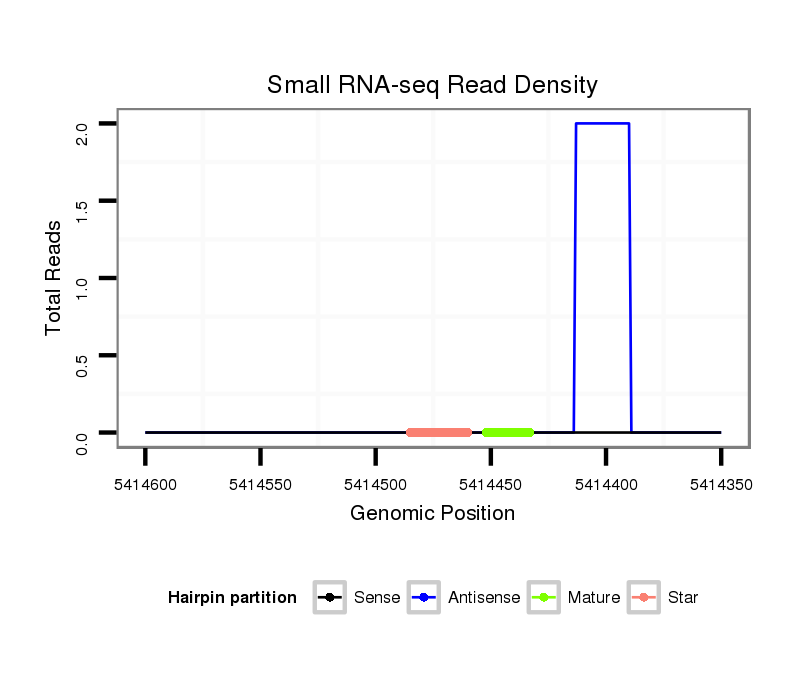
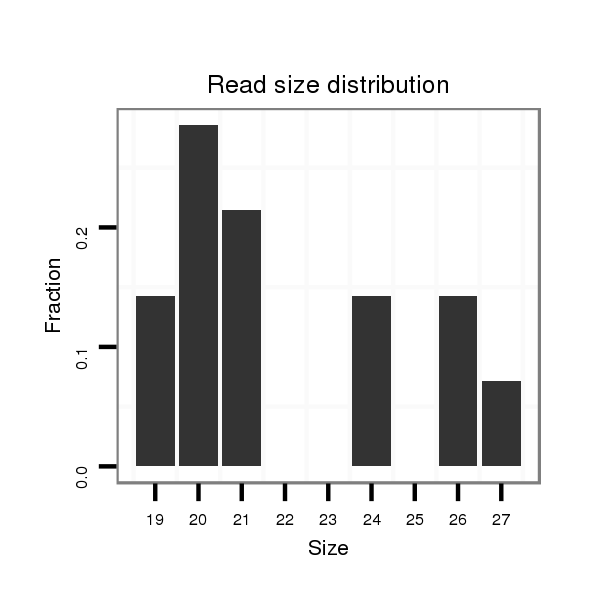
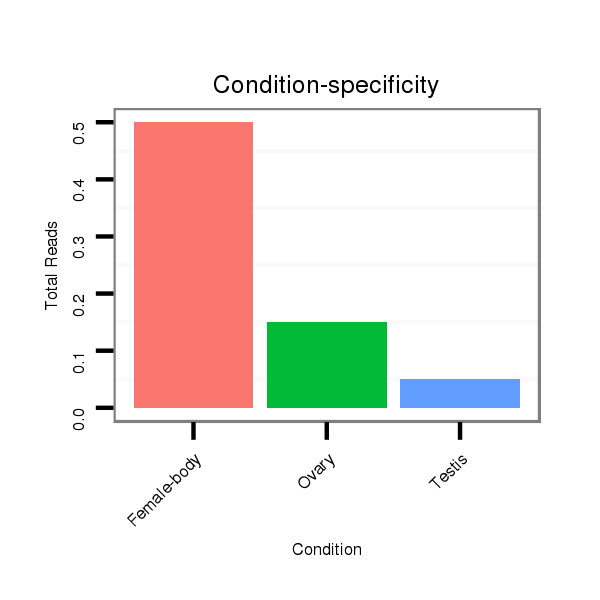
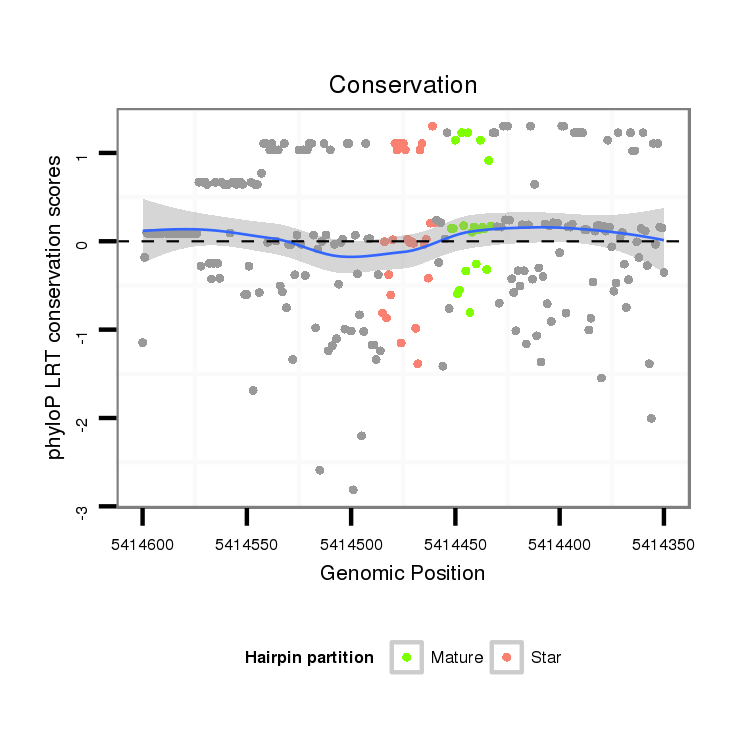
ID:dps_3331 |
Coordinate:XR_group8:5414400-5414550 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -7.4 | -6.9 |
 |
 |
CDS [Dpse\GA28278-cds]; exon [dpse_GLEANR_7844:2]; intron [Dpse\GA28278-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TATCCACTCCAAGCGCCTGACTGGCAGATATCGCGACATGCCAATCTTCGGTAAGCTATATCTCAGACAAATAGGGCCAGATCGGTGAAGGACCTATTTTCCCAGAATCGCGAGGACCCAGACTGTCGATTTGCACCAAAATCTCGACTTCGAAAATGAGGCCGATTTTTGGCAAATTTAATGATGTAACCATCATGATTTTTTGCGAAAATCGTGAAAAAATGGATGTCCTTCTTTCCGATGGCAGTCGA *******************************************************************************************************************....((...((((....((..((.....(((....)))...))..))))))))*********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902010 ovaries |
SRR902011 testis |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................TTCGAAAATGAGGCCGATTT................................................................................... | 20 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 |
| .........................................................................................................................................................................................................TTTGCGAAAATCGTGAAAAAATG........................... | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| ..............................................................TCAGACAAGTAGGGCCAGATCGGTG.................................................................................................................................................................... | 25 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| ...................................................................................................................ACCCAGACTGTCGATTTGCACCAAAA.............................................................................................................. | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| ........................................................................................................................................................AAAATGAGGCCGATTTTTGGC.............................................................................. | 21 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| .........................................................ATATCTCAGACAAATAGGGC.............................................................................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .......................................................................................................................................................GAAAATGAGGCCGATTTTTGG............................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .....................................................AGCTATATCTCAGACAAATAGGGC.............................................................................................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..............................................................TCAGACAAATAGGGCCAGATCGGT..................................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| .......................................................................TAGGGCCAGATCGGTGAAG................................................................................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................GATGTCCTTCTTTCCGATGGC...... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .......................................................................................................AGAATCGCGAGGACTCAGACTATCGACT........................................................................................................................ | 28 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................................................TTTTGGCAAATTTAATGATGTAACCAT.......................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................................CGAAAATGAGGCCGATTTT.................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
ATAGGTGAGGTTCGCGGACTGACCGTCTATAGCGCTGTACGGTTAGAAGCCATTCGATATAGAGTCTGTTTATCCCGGTCTAGCCACTTCCTGGATAAAAGGGTCTTAGCGCTCCTGGGTCTGACAGCTAAACGTGGTTTTAGAGCTGAAGCTTTTACTCCGGCTAAAAACCGTTTAAATTACTACATTGGTAGTACTAAAAAACGCTTTTAGCACTTTTTTACCTACAGGAAGAAAGGCTACCGTCAGCT
***********************************************************************************....((...((((....((..((.....(((....)))...))..))))))))******************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M062 head |
SRR902010 ovaries |
SRR902011 testis |
M059 embryo |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................TTGGTAGTACTAAAAAACGCTTTT........................................ | 24 | 0 | 20 | 2.30 | 46 | 46 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................GGTTTTAGAGCTGAAGCTTTTACT............................................................................................ | 24 | 0 | 20 | 0.50 | 10 | 3 | 0 | 6 | 1 | 0 | 0 |
| ...........................................................................................................................ATAACTAAACGTGTTTTTAGA........................................................................................................... | 21 | 3 | 20 | 0.30 | 6 | 0 | 6 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AACTAAACGTGTTTTTAG............................................................................................................ | 18 | 2 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................ACTAAACGTGTTTTTAGA........................................................................................................... | 18 | 2 | 20 | 0.15 | 3 | 2 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................AACTAAACGTGTTTTTAGA........................................................................................................... | 19 | 2 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................................GGGTCTGACAGCTAAACGTGGTTTT.............................................................................................................. | 25 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ........................................................................................................................................................................................ACATTGGTAGTACTAAAAAACGCTT.......................................... | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CCGGCTAAAAACCGTTTAAATT...................................................................... | 22 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................TTAGAAGCCATTCGATATAGAGT.......................................................................................................................................................................................... | 23 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........TCGCGGGCTGACCGTGT............................................................................................................................................................................................................................... | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................TACATTGGTAGTACTAAAAAACGCTT.......................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GTTTTAGAGCTGAAGCTTTTACT............................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................CGGTCTAGCCACTTCCTGGAT........................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TCTGTTTATCCCGGTCTAGC....................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CCGGCTAAAAACCGTTTAAATTA..................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................CTCCTGGGTCTGACAGCTA......................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CTGACAGCTAAACGTGGTTTTAGAGCT........................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................CCCGGTCTAGCCACTTCCTGGA............................................................................................................................................................ | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................TTTAGAGCTGAAGCTTTTACT............................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AGCTAAACGTGGTTTTAG............................................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AGCTAAACGTGGTTTTAGAG.......................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................GGTCTGACAGCTAAACGTGG.................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CTGTTTATCCCGGTCTAGCCACTTCC................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................CTAAACGTGCTTTTAGAA.......................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................CGTGGTTTTAGAGCTGAAGCTTTTACT............................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CTGAACGTGGTTTTAGAG.......................................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TGGTTTTAGAGCTGAAGCTTTTACT............................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AGCTAAACGTGGTTTTAGAGCTGAAGCT.................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AGAGCTGAAGCTTTTACTCCG......................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............GCGGACTGAGGGCCTATA............................................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................ACAACTAAACGTGTTTTTAG............................................................................................................ | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CGTGGTTTTAGAGCTGAAGCT.................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................TGTTTATCCCGGTCTAGCCACTT.................................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................CGGTCTAGCCACTTCCTGGA............................................................................................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group8:5414350-5414600 - | dps_3331 | TATCCACTCCAAGCGCCTGACTGGCAGATATCGCGACATGCCAATCTTCGGTAAGCTATATCTCAGACAAATAGGGCCAGATCGGTGAAGGACCTATTTTCCCAGAATCGCGAGGACCCAGACTGTCGATTTGCACCAAAAT-CTCGACTTCGAAAATGAGGCCGATTTTTGGCAAA-TTTAA--TGATGTAACCATCA----------------------------------TGATTTTTTGCGAAAATCGTGAAAAAATGGATGTCC-TTCTTTCCGATGGCAGTCGA |
| droPer2 | scaffold_38:272166-272416 + | dpe_1536 | TATCCACTCCAAGCGCCTGACTGGCAGATATCGCGACATGCCAATCTTCGGTAGGCTATATCTCAGACAAATAGGGCCAGATCGGCGAAGGACCTACTTTCGCAGGATCGCGAGGACCCAGACTGTCGATTTGCACCAAAAT-CTCGACTTCGAAAATGAGGCCGATTTTTGGCAAA-TTTAA--TGATGTAACCATCA----------------------------------TGATTTTTTGCGAAAATCGTGAAAATATGGATGTCC-TTCTTTCCGATGGGAGTCGA |
| droVir3 | scaffold_13049:20713481-20713675 - | A---------------------------------------------------------TATCTCGGGTATTACAAGTCTGATTTCCGAGCGGTATACCTTGTTTTACTCGTACAAACCTGGACTATCGATTGCCATCGAAAAATTTCAATTCAAAGTTTTAACCAAA-ATTGACAGA-AATGGCAAGGGGTT-ACATCA----------------------------------CAATT-TTGGCGAAAATCGGCTCGGTTGGAAAATCAACAATTTCTGATGCCAATCTG | |
| droAna3 | scaffold_13334:477995-478220 + | AA-------------------------AAATCAAAATCTGCC-ATCTTCCCAAAGCTATAACTCGGCTAATATTGGTCCGATCGGTGAATGTTATACCTTCCCGATCTTGTATTTTGGAGTACTCTCATTCCCCATCCAAAT-CTGGAAAACTTAAATCTGACCGGATTTTCACAAA-TTCTGCAAGGGGTA-AGGTCA----------------------------------CAAAAATTGCCGAAAATCGACAAAAAATGGTGGTGC-CATTTTTTAATGCAAATCGA | |
| droBip1 | scf7180000396425:357592-357762 + | C-------------------------------------------------------CATATCTCGGCCATTAGTGGTCCGATCTGGCAACGCTTGGCATTTTCGGAATCAGCGTCCCAAAGACTGTCGAATTGCATACCAAT-TTT-------------------------GTCAAAATCAGACAAAAAATT-TTTCCGCGAAAATTGGAAAAAATCGTGATGTATAGCCTTTCCATT-TTTGCG------------------------------------AAAAATCGG | |
| droKik1 | scf7180000302686:1407937-1408044 + | CT---------------------------------------------------------------------------------------------------------------------------------------AAATT-CAGGATTTCACAATTTTGGTCGATTTTTGAAAAA-TTTAA--CGATGTAACCCCTT---------------------------------TCATTTTTTTGAAAAAATTACTGAAAAATGGTTTTCC-TCCT----GACA-----AGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/18/2015 at 06:39 AM