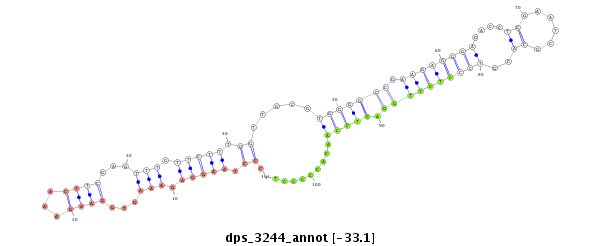
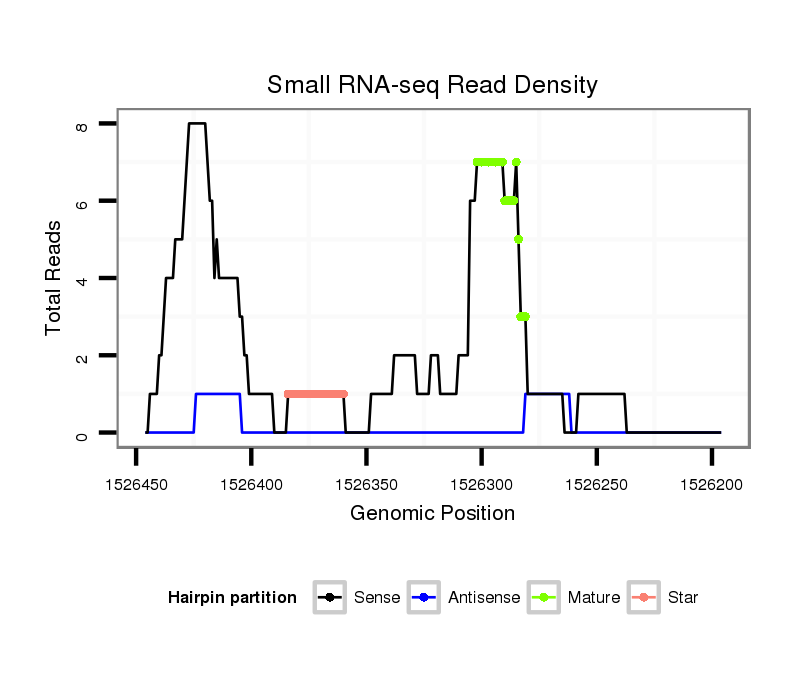
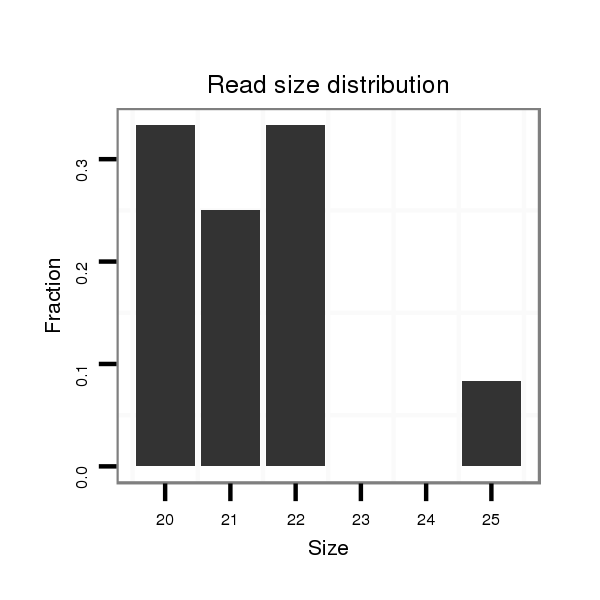
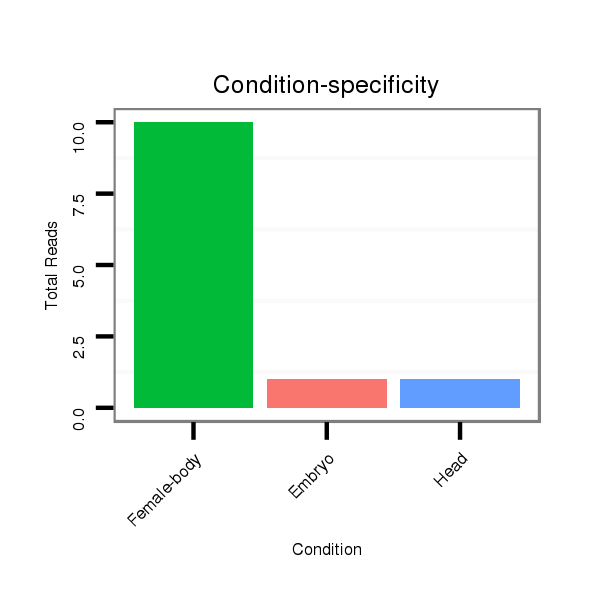
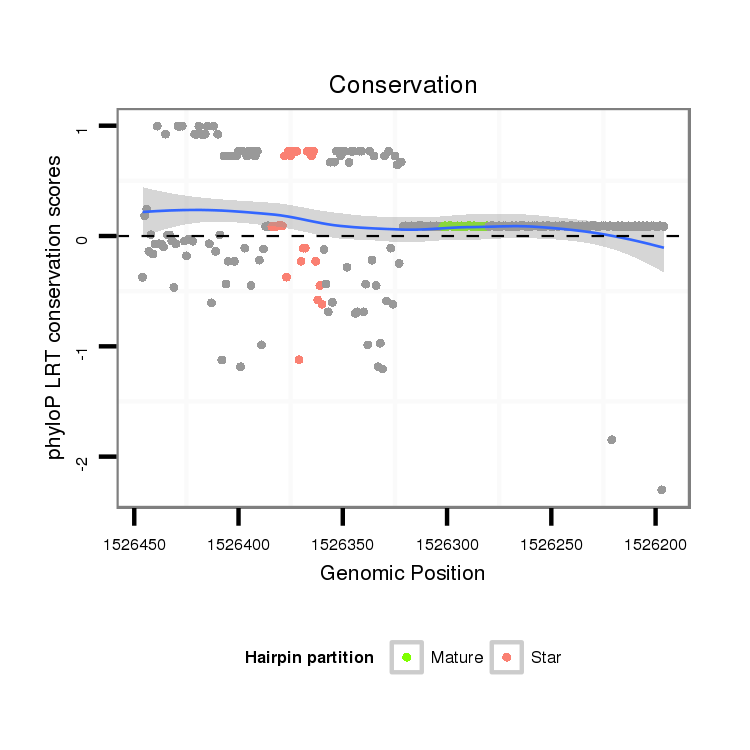
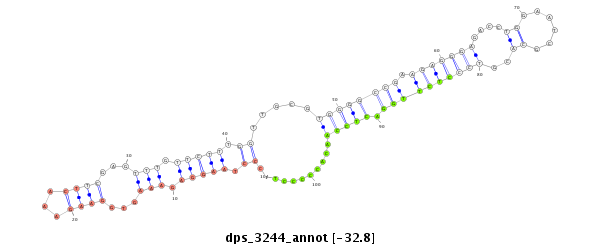
ID:dps_3244 |
Coordinate:XR_group8:1526246-1526396 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -32.8 | -32.8 | -32.7 |
 |
 |
 |
exon [dpse_GLEANR_8092:1]; CDS [Dpse\GA20886-cds]; intron [Dpse\GA20886-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATTGCACTAGTGATAGTTTTTCTCGGCTGCCTTCTCGTGGCACCACTGAGGTGAGTCCTGTGCCCTAAGGAGAAAGTGGAAGAAACTTCGAGTTTGTTCTTTGGTTGCGTGGGGCCGAAGAGGGAGACCTGGAATCGCACGTCCCTCTTGGACTCCAACACCCCCTCAAGGACAAGCACTCAATGTCAATGGTGAACCAGTTTCGATGGAGTCACAATCCTTTGTTGCACACCTGCACACACTGTAGGAGC **************************************************************.((.(((((.(((...((((...))))...))).))))).)).....(((((((.((((((((....((......))..)))))))))).))))).........************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M062 head |
M022 male body |
M059 embryo |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................CTCTTGGACTCCAACACCCCCT..................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TCCCTCTTGGACTCCAACACC......................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................TCCCTCTTGGACTCCAACACCC........................................................................................ | 22 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ......................TCGGCTGCCTTCTCGTGGCACA............................................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................ATGGTGAACCAGTTTCGATGG.......................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................TCTTGGACTCCAACACCCCCTT.................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........GTGATAGTTTTTCTCGGCTGC............................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................TTCTCGTGGCACCACTGAGGTGAGT................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..TGCACTAGTGATAGTTTTTCTCGGC................................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................CCCTAAGGAGAAAGTGGAAGAAACT.................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................GTGGGGCCGAAGAGGGAGAC........................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......CTAGTGATAGTTTTTCTCGGCT............................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................TTTTCTCGGCTGCCTTCTCGTGGCAC................................................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........AGTGATAGTTTTTCTCGGCTGC............................................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................AGACCTGGAATCGCACGTCC........................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................CTTTGGTTGCGTGGGGCCGA..................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GCACGTCCCTCTTGGACTCC............................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CGTCCCTCTTGGACTCCAACACA......................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................CCCCTCAAGGACAAGCACTCA..................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................TTTCTCGGCTGCCTTCTCGTGGCACCA.............................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............TAGTTTTTCTCGGCTGCCT........................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................TTCTCGGCTGCCTTCTCGTGGC.................................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................AATGTGAATGGTGAACCA.................................................... | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................GTTCTTTGATGGCGTGGGG......................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GTCACTCCCCTTTGTTGCAC..................... | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................AATCTCGCTGGTGAACCAG................................................... | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................TTTGGTGGCGTGGGGCAG...................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
TAACGTGATCACTATCAAAAAGAGCCGACGGAAGAGCACCGTGGTGACTCCACTCAGGACACGGGATTCCTCTTTCACCTTCTTTGAAGCTCAAACAAGAAACCAACGCACCCCGGCTTCTCCCTCTGGACCTTAGCGTGCAGGGAGAACCTGAGGTTGTGGGGGAGTTCCTGTTCGTGAGTTACAGTTACCACTTGGTCAAAGCTACCTCAGTGTTAGGAAACAACGTGTGGACGTGTGTGACATCCTCG
*************************************************************************************.((.(((((.(((...((((...))))...))).))))).)).....(((((((.((((((((....((......))..)))))))))).))))).........************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM444067 head |
V112 male body |
M062 head |
M059 embryo |
GSM343916 embryo |
M022 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................CCCTCTGGACCTAAGCTTG............................................................................................................... | 19 | 2 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GTGTGGACGTGTATGACATCC... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................TGTTGTGGGGGAGTTCCTGT............................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................AGTTCCTGTTCGTGAGTTAC.................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................AGCCGACGGAAGAGCACCGT................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..ACGTGGTCACTATCAAAGAGA.................................................................................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CCCTCTGGACCTAAGCTTGG.............................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................GTTCCTTTTCGGGAGCTACAGT............................................................... | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................CAACGCAGGAGCACCGTGGT.............................................................................................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................ACCCTCTGGACCTAAGCTTG............................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CTGGACCGTACCGTGCAG............................................................................................................ | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................CCTCTGGACCTAAGCTTGG.............................................................................................................. | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................GGGGGAGTTGGTGTTCGT......................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................CCTCAGTGTTAGGCCACG.......................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group8:1526196-1526446 - | dps_3244 | ATTGCACTAGTGATAGTTTTTCTCGGCTGCCTTCTCGTGGCA---------CCACTGAGGTGAGTCCTGTGCCCTAAGGAGAAAGTGGAAGAA---ACTTCGAGTTTGTT---CTTTGGTTGCGTGGGGCCGAAGAGGGAGACCTGGAATCGCACGTCCCTCTTGGACTCCAACACCCCC------TCAAGGACAAGCACTCAATGTCAATGGTGAACCAGTTTCGATGGAGTCACAATCCTTTGTTGCACACCTGCACACACTGTAGGAGC |
| droPer2 | scaffold_24:1527514-1527769 + | ATTGCACTAGTGATAGTTTTTCTCGGCTGCCTTCTCGTGGCA---------CCACTGAGGTGAGTCCTGTGCCCTAAGGAGAAAGTGGAAGAA---ACTTCGAGTTTGTT----TTTGGTTGCGTGGGGCTGAAGAGGGAGACCTGGAATCGCACGTCCCTCTTGGACTCCAACACCCCCACCTCATCAAGGACAAGCACTCAATGTCAATGGTGAACCAGTTTCGATGGAGTCACAATCCTTTGTCGCACACCTGCACACACTGTAGGACC | |
| droWil2 | scf2_1100000004768:2767499-2767501 + | AGA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13337:16054302-16054426 - | TTTCTCCTGGTGGCCTCTTTGCTTTGCTGCCTCTTGGCCGCCAGCTCCAGTCCCCTAAAGTAAGTAGC---------GCAGAAATGAAAAGACGCAATTACG---TTGTTTTGTTTTTGTTGCCTTGAAATGGAAAG--------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396589:379312-379434 + | TTTCTCCTGGTGGCCTCTTTGCTTTGCTGCCTCTTGGCTGTCAGCTCCAATCCCCTCAAGTGAGTACC---------GCAGAAACGAAAAGAC---GCAATTACTTTGTT-----TTGTTTTTGTTGGCTTGAAATGGAA------------------------------------------------------------------------------------------------------------------------------------ | |
| droKik1 | scf7180000302274:301866-301903 + | CTTCTAATATGGGCCATTTTGGCCTGCTGCCTTGTGGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/18/2015 at 06:24 AM