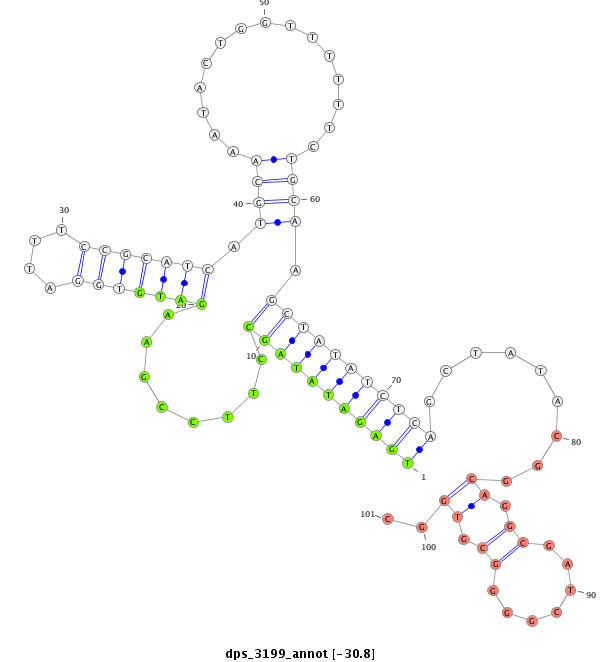

ID:dps_3199 |
Coordinate:XR_group6:13164537-13164687 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -29.9 |
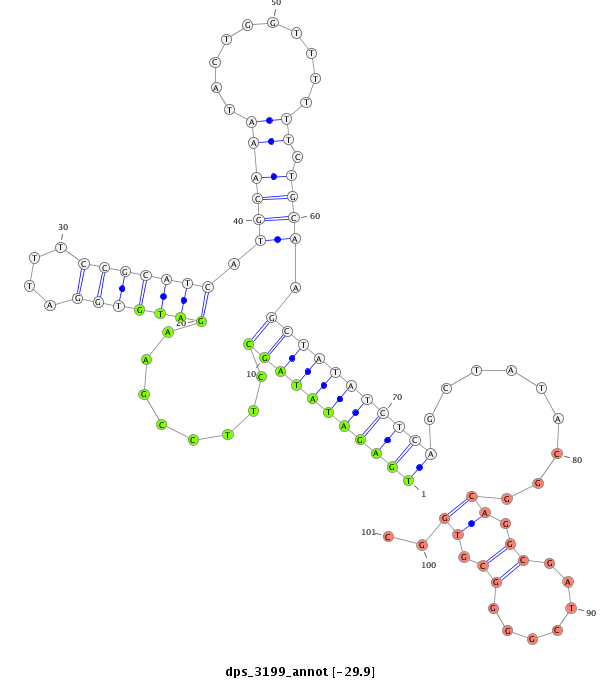 |
CDS [Dpse\GA23963-cds]; exon [dpse_GLEANR_13495:1]; intron [Dpse\GA23963-in]
No Repeatable elements found
| -----------#######################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TCGAGATTTTGATGCCAATCGATAAGGTTAATCCCCACTGTCCTGGGAAGGTAGGTCCTTCGCCGAATTGGCCCTATTTGCCTGAGATATAGCCTTCCGAAGATGTGGATTTCCGCATCATGCAAATACTGGTTTTTTCTGCAAGCTATATCTCAGCTATACGGCAGGCGATCGGGGCGTGGCATGTCATTCCGTGACCGGGAGAGTCCTGCCAACCGATTGGCACCAGAAAAAAGGATATCCATTCCATC **********************************************************************************(((((((((((........(((((((....))))))).((((...............)))).))))))))))).........((.((.......)).))..******************************************************************** |
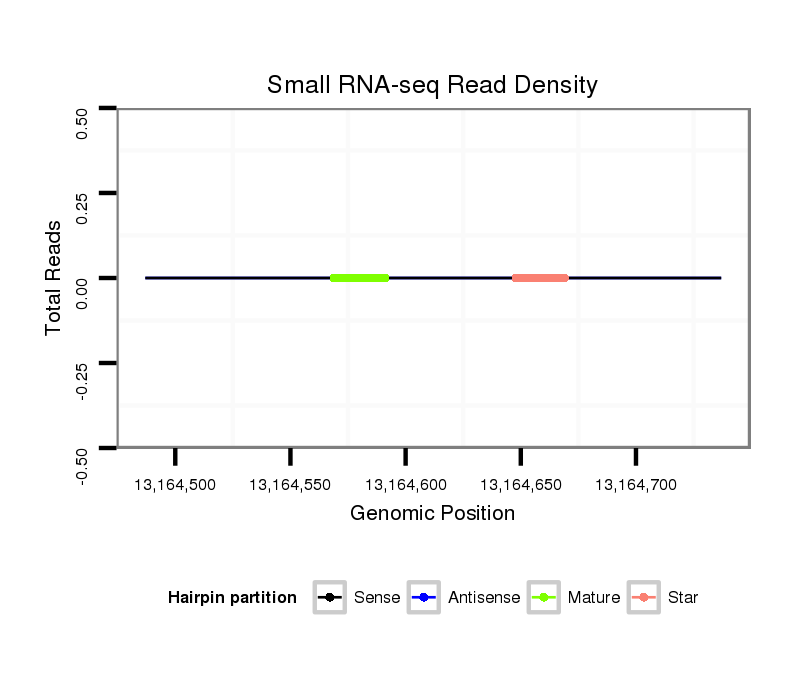
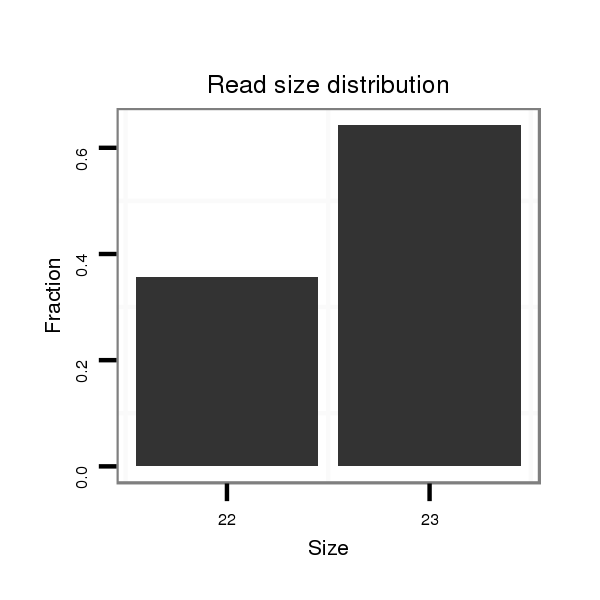
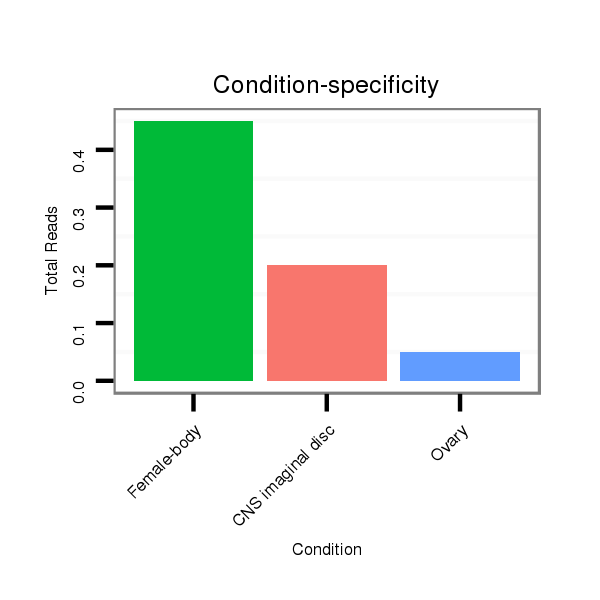
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902012 CNS imaginal disc |
GSM343916 embryo |
SRR902010 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................TGAGATATAGCCTTCCGAAGATG.................................................................................................................................................. | 23 | 0 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 |
| ...........................................................TCGCCGAAATGCCCCTATTGG........................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................CGGCAGGCGATCGGGGCGTGGC.................................................................... | 22 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................GCACATCCAGGTTTTTTCTGC............................................................................................................. | 21 | 3 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................TATCTCTGCTATACGGCAGGCGA................................................................................ | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................ATTGGTTTTTGCTGCAAGC......................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................GAGATATAGCCTTCCGAAGATG.................................................................................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................GTCCTGCCAACCGATTGGCA.......................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TGCCAACCGATTGGCACCAGAAA................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................TCCTGCCAACCGATTGGCAC......................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GGTCCTGCCAACCGATTGGCACCAGA..................... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................GTCCTGCCAACCGATTGGCACCAGAT.................... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................TATAGCCTTCCGAAGATGTCGACT............................................................................................................................................ | 24 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................TGAGATATAGCCCTCCGAGGCTGT................................................................................................................................................. | 24 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................CTGAGATATAGCCTTCCGAAGAT................................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................................................................TCCTGCCAACCGATTGGCACCA....................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
AGCTCTAAAACTACGGTTAGCTATTCCAATTAGGGGTGACAGGACCCTTCCATCCAGGAAGCGGCTTAACCGGGATAAACGGACTCTATATCGGAAGGCTTCTACACCTAAAGGCGTAGTACGTTTATGACCAAAAAAGACGTTCGATATAGAGTCGATATGCCGTCCGCTAGCCCCGCACCGTACAGTAAGGCACTGGCCCTCTCAGGACGGTTGGCTAACCGTGGTCTTTTTTCCTATAGGTAAGGTAG
********************************************************************(((((((((((........(((((((....))))))).((((...............)))).))))))))))).........((.((.......)).))..********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
M059 embryo |
V112 male body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .......................................................................GGGATAAACGGACTCTATATC............................................................................................................................................................... | 21 | 0 | 20 | 0.55 | 11 | 11 | 0 | 0 | 0 |
| ...........................................................AGCGGCTTTACGGGGATAACC........................................................................................................................................................................... | 21 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................................................CACCGTACAGTAAGGCACTGGCCCTCT.............................................. | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................AGCGGCTTTACGGGGATAAC............................................................................................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................................................................AGTACGTTTATGACCAAAAAAGAC.............................................................................................................. | 24 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................AGCGGCTTTACGGGGGTAAAC........................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................CCGGGATAAACGGACTCTATATC............................................................................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| ..........................................................AAGCGGCTTTACGGGGGTAA............................................................................................................................................................................. | 20 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................AGCGGCTTTACAGGGGTAA............................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGGCTAACCGTGGTCTTTTTTCCTAT........... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................CGGGATAAACGGACTCTATATC............................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................TGTCGGAAGGGATCTACACC............................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group6:13164487-13164737 + | dps_3199 | TCGAGATTTTGAT-----------GCCAA-TCGATAAGGTTAA--TCCCCACTGTCC-TGGGAAGGTAGGTCCTTCGCCGAATTG-GCCCT-AT-TTGCCTGAGATAT----------------------------------------AGCCTTCCGAA----------------------------GATGTGGATTTCCGC--ATCATGCAAA---TACTGGTTTTTTCTGC-AAGCTATATCTCAGCTATACGGCAGGCGATCGGGGCGTGGCATGTCATTCCGTGACCGGGAGAGTCCTGCCA------------------------------------------ACCGATTGG---------CACC-------------AGAAAA----AAGGATATCCATTCCATC |
| droPer2 | scaffold_88:202597-202847 + | TCGAGATTTTGAT-----------GCCAA-TCGACAGGGTTAA--TCCCCACTGTCC-TGGGAAGGTAGGTCCTTCGCCGAACTA-GCCCT-AT-TTGCCTGAGATAT----------------------------------------AGCCTTCCGAA----------------------------GATGTGGATTTCCGC--ATCATGCAAA---TACTGGTTTTTTCTTT-AAGCTATATCTCAGCTATCCGGAAGGCGATCGGGGCGTGGCATGTCATTCCGTGACCGGAAGAGTCCTGCCA------------------------------------------ACCGATTGG---------CACC-------------AGAAAA----AAGGATATCCATTCCATC | |
| droVir3 | scaffold_12875:16314946-16315229 + | TTTTGA------T-----------GCCAA-TCGATAGAACTGA--TCCTTACGAGTC-AAAAATGGTATGAAATTTCA-AAATCGGACGTT-AT-TTGCCCGAGATAT----------------------------------------GCCA---AAAAATCATAAAAAATGGTTGATTTTGAAAACGAGTTGGTTTTCCGTCAATCCCATCGA---TATTGAATTTCTTTGA-ATGAGAATTATTAGTAGGGATCCGTACGAATTTAGAAAGGTATAACATTTCAAAATCAAACGATATTTACCGGAGATATACAATAAAAATCATCGAAAAAGTTTTGATTTTTG--------------------------------------------------------ACCCGACC | |
| droAna3 | scaffold_13117:1877905-1878183 - | GCCCGATTTTCACAATCCTGGGATGCAGT-TCGAGGGATATCGGAGC---CCTGATCGCAAAATGAC-ATCCCTTTTCCAAATCC-GTCCAGAA-ATAGCCAAGATATGCGCTCCGAA-----AGTTCAGCCTTGGCGGCCGAAACGCCGGCCGC-------------------------------------------------CG------------CTTGGGCATCTTGCCAAGCTCATATCTTGGCCATTTCCCAACCGATCTTGAAAAAGGAAGCCATTTTGGAATCAGGATCTCAATGTCC------------------------------------------TTCGATCTGCATCTAAAGTTTT-------------TCAAAA----TCGGACAATAAAATTTTC | |
| droBip1 | scf7180000396544:429067-429356 - | TCTAGATTTTGAT-----------ACAGAATTTAAGATATTTT--TACCAGGAGTAT-TTTGATGT--GTTCCTTAAAGAAATCA-GACGT-ATACTTTTTAAAATAT----------------------------------------GTGTTTTAGTAGACACAAAAATTTCATGTTTTTTATTAC--GTTGGTACTTTTTTATTTTCGAAAAATTTTTC---GAACTTTGGAAGATCATATCTCAGTTATTTGGTGGTCGATCCAAGAATGTTATATCATTTCGATCTCAGAA------------------------------------AAAGTTGTACTTTTCAATACGACTTT---------CTCT-------------AGTAAATTATAAAAATTTAAAATTTTTT | |
| droKik1 | scf7180000302351:865569-865805 - | TTTAGTTAAGTAT-----------GCAGA-CTTATTAACCTTA--AAAAAA--------------------------------------------CTAACTCAGAACTGCTTTCCGAATTGAATTTAATGCATTTTTGGCTGAGTTATAGATTTTTTAA----------------------------GATTTGATTTTCCCC--AA-----AAA---TATC----------GT-AATCTAAAACTCAGTGA---AGGAGTCATTTCCGACCCTATAAACCATATATATTCTTGATCAGCATCGACA------------------------------------------GCCGAGTAA---------TATAAGCCATGTTTTGAAGAAAA----AAAA---------ATTTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/18/2015 at 06:24 AM