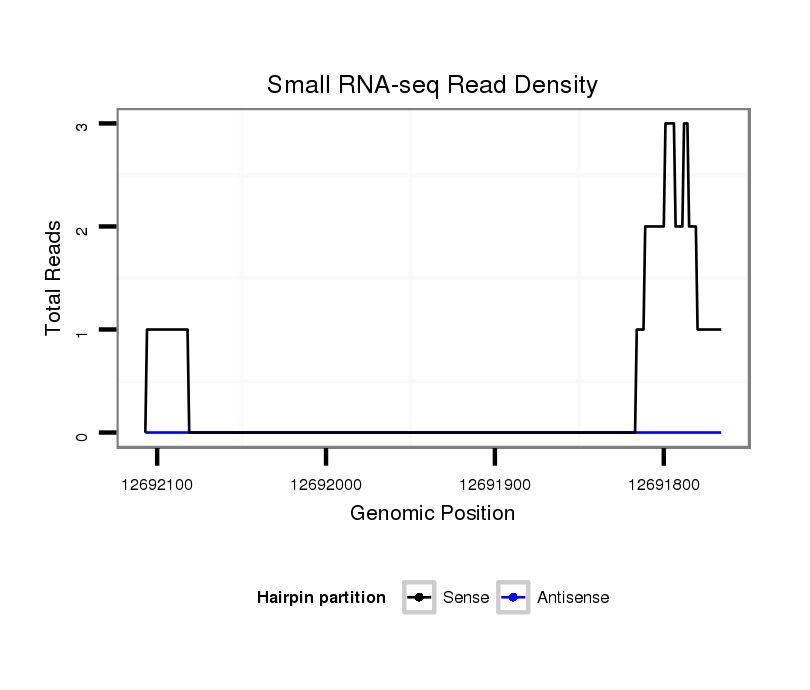
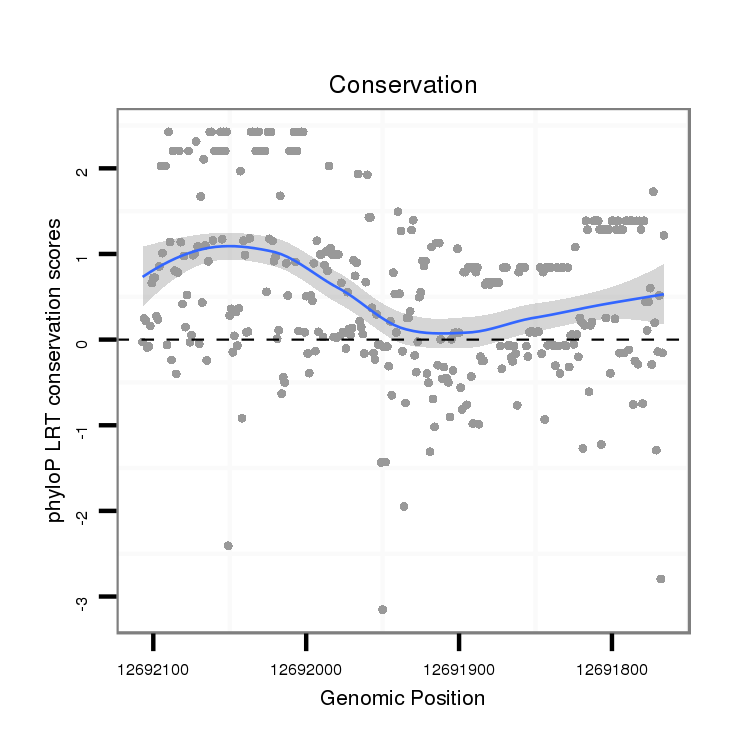
ID:dps_3187 |
Coordinate:XR_group6:12691816-12692057 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dpse_GLEANR_12039:1]; CDS [Dpse\GA11947-cds]; CDS [Dpse\GA11947-cds]; exon [dpse_GLEANR_12039:2]; intron [Dpse\GA11947-in]; Antisense to intron [Dpse\GA16901-in]
No Repeatable elements found
| --------##########################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CAATTGAAATGGCAGCCATGGCAGCAGCAACAGTCAATAACCACAAAGGGGTAAGTCTGACGCGTCTGCTTTAACTTGGCCTTTAAGCCGCTCCGAGGGTTGGTTTACGGGACATTGCTTATGACACACGCGGTCGCCAGACAAATACGCGGCCAGTGTGGCACATGCATTAGTGTGTGTGTATGTATTTGCAAACATGCATATTTTATTGCATTGCTCAGGGAGATGTTTTTAATTAACGGGTGCGTAAATCGATCGAAAGGGTGATCCAAACTTCAATTTCTTTCCGCAGCCCAAATCCCGTCGCAACGAGCGCAACATATTGACCTTCTACGAGAAGAT ***********************************..........(((((.(((((.(((.((....)).)))))))).)))))(((((((((...))).))))))((((((((((.((..((((((((..((((((...........)))))).)))))(((.(((((..(((((((((((.(((....))).)))))))))))....))))))))))).)).))))............(((((((..........((((((((((........)))..))))))))))).)))...))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M022 male body |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................CGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 18 | 2 | 3 | 22.33 | 67 | 60 | 6 | 1 |
| ............................................................................................CGGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 19 | 3 | 10 | 20.00 | 200 | 173 | 27 | 0 |
| ...........................................................................................TCCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 20 | 2 | 1 | 18.00 | 18 | 13 | 5 | 0 |
| ...........................................................................................TCCGAGAGTTGGTTTAAGGGG...................................................................................................................................................................................................................................... | 21 | 3 | 1 | 10.00 | 10 | 7 | 3 | 0 |
| ..........................................................................................CTCTGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 21 | 3 | 2 | 9.50 | 19 | 13 | 6 | 0 |
| ..........................................................................................CTCCGAGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 20 | 2 | 1 | 7.00 | 7 | 3 | 3 | 1 |
| ...........................................................................................TCCGAGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 19 | 2 | 2 | 6.50 | 13 | 9 | 4 | 0 |
| ..........................................................................................CTCGGAGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 20 | 3 | 2 | 6.00 | 12 | 7 | 5 | 0 |
| ..........................................................................................CTCGGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 21 | 3 | 1 | 6.00 | 6 | 3 | 3 | 0 |
| ..........................................................................................CTCAGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 21 | 3 | 2 | 6.00 | 12 | 9 | 3 | 0 |
| ..........................................................................................CTCCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 21 | 2 | 1 | 4.00 | 4 | 3 | 1 | 0 |
| ............................................................................................TCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 19 | 3 | 13 | 3.69 | 48 | 36 | 12 | 0 |
| ...........................................................................................TCAGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 20 | 3 | 6 | 3.33 | 20 | 12 | 8 | 0 |
| ............................................................................................CCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 19 | 2 | 1 | 3.00 | 3 | 2 | 1 | 0 |
| ...........................................................................................TCGGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 20 | 3 | 4 | 3.00 | 12 | 10 | 2 | 0 |
| .........................................................................................TCTCCGAGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 21 | 3 | 3 | 2.00 | 6 | 4 | 0 | 2 |
| ..........................................................................................CTCAGAGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 20 | 3 | 4 | 1.75 | 7 | 7 | 0 | 0 |
| ............................................................................................CCGAGAGTTGGTTTAAGGGG...................................................................................................................................................................................................................................... | 20 | 3 | 2 | 1.50 | 3 | 2 | 1 | 0 |
| ..............................................................................................GAGAGTTGGTTTAAGGGA...................................................................................................................................................................................................................................... | 18 | 2 | 8 | 1.13 | 9 | 9 | 0 | 0 |
| .........................................................................................TCTCCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................................CTTCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................GCCCAAATCCCGTCGCAACGAGC............................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .AATTGAAATGGCAGCCATGGCAGCA............................................................................................................................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................................CTCCCAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ........................................................................................CTCTCCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................ATATTGACCTTCTACGAGAAGAT | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................................CCCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 20 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................AATCCCGTCGCAACGAGCGCAACATA.................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................ACGAGCGCAACATATTGAC............... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................................TCCCAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.67 | 4 | 4 | 0 | 0 |
| ............................................................................................CCGAGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 18 | 2 | 3 | 0.67 | 2 | 2 | 0 | 0 |
| ............................................................................................CGGAGGGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ...........................................................................................TCCGAGAGTTGGTTTAAGGGT...................................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................CGCAACGAGCGCAACCCTT................... | 19 | 3 | 20 | 0.45 | 9 | 9 | 0 | 0 |
| ...........................................................................................TTCGAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .......................................................................................ACTCTCCGAGAGTTGGTTTA........................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ............................................................................................CCGAGAGTTGGTTTAAGGGT...................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ..............................................................................................GAGGGTTGGTTTAAGGGG...................................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.25 | 2 | 2 | 0 | 0 |
| .............................................................................................CGAGAGTTGGTTTAAGGGT...................................................................................................................................................................................................................................... | 19 | 3 | 19 | 0.21 | 4 | 2 | 1 | 1 |
| ...............................................................................................AGAGTTGGTTTAAGGGTC..................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.20 | 4 | 4 | 0 | 0 |
| .............................................................................................GGAGGGTTGGTTTAGGGG....................................................................................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ..........................................................................................CTCCCAGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 |
| ..........................................................................................CTCTCAGGGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 |
| ............................................................................................CCCAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 19 | 3 | 19 | 0.11 | 2 | 2 | 0 | 0 |
| .............................................................................................CGAGAGTTGGTTTAAGGTA...................................................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 |
| ...........................................................................................TCCGTGAGTTGGTTTAAGG........................................................................................................................................................................................................................................ | 19 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 |
| ............................................................................................CCTAGAGTTGGTTTAAGGG....................................................................................................................................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 |
| ..........................................................................................CTCAGAGAGTTGGTTTAAG......................................................................................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 |
|
GTTAACTTTACCGTCGGTACCGTCGTCGTTGTCAGTTATTGGTGTTTCCCCATTCAGACTGCGCAGACGAAATTGAACCGGAAATTCGGCGAGGCTCCCAACCAAATGCCCTGTAACGAATACTGTGTGCGCCAGCGGTCTGTTTATGCGCCGGTCACACCGTGTACGTAATCACACACACATACATAAACGTTTGTACGTATAAAATAACGTAACGAGTCCCTCTACAAAAATTAATTGCCCACGCATTTAGCTAGCTTTCCCACTAGGTTTGAAGTTAAAGAAAGGCGTCGGGTTTAGGGCAGCGTTGCTCGCGTTGTATAACTGGAAGATGCTCTTCTA
***********************************..........(((((.(((((.(((.((....)).)))))))).)))))(((((((((...))).))))))((((((((((.((..((((((((..((((((...........)))))).)))))(((.(((((..(((((((((((.(((....))).)))))))))))....))))))))))).)).))))............(((((((..........((((((((((........)))..))))))))))).)))...))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M022 male body |
M059 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................GAGGCTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 21 | 2 | 1 | 8.00 | 8 | 8 | 0 | 0 | 0 |
| ..........................................................................................GAGCCTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 21 | 3 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 |
| .........................................................................................AGAGGCTCTCAACCAAATTCC........................................................................................................................................................................................................................................ | 21 | 3 | 3 | 7.00 | 21 | 14 | 6 | 1 | 0 |
| ...........................................................................................AGGCTCTCAACCAAATTCCCC...................................................................................................................................................................................................................................... | 21 | 3 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| ........................................................................................GAGAGGCTCTCAACCAAATTC......................................................................................................................................................................................................................................... | 21 | 3 | 3 | 3.67 | 11 | 6 | 4 | 1 | 0 |
| ..........................................................................................GAGGGTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 21 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| .........................................................................................TGAGGCTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 22 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..........................................................................................GAGACTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 21 | 3 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 |
| .......................................................................................TGAGAGGCTCTCAACCAAAT........................................................................................................................................................................................................................................... | 20 | 3 | 4 | 1.25 | 5 | 4 | 1 | 0 | 0 |
| .........................................................................................CGAGCCTCTCAACCAAATTCC........................................................................................................................................................................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................GAGAGGCTCTCAACCAAATT.......................................................................................................................................................................................................................................... | 20 | 3 | 5 | 1.00 | 5 | 4 | 0 | 1 | 0 |
| ..........................................................................................GAGTCTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................GGCTCTCAACCAAATTCCCC...................................................................................................................................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................AGCCTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 |
| ..........................................................................................GAGCCTCTCAACCAAATTCC........................................................................................................................................................................................................................................ | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................AGAGGCTCTCAACCAAATTC......................................................................................................................................................................................................................................... | 20 | 3 | 9 | 0.44 | 4 | 2 | 0 | 2 | 0 |
| ............................................................................................GCCTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.40 | 4 | 4 | 0 | 0 | 0 |
| ..........................................................................................GAGGGTCTCAACCAAATTCC........................................................................................................................................................................................................................................ | 20 | 3 | 8 | 0.38 | 3 | 3 | 0 | 0 | 0 |
| ................GTACAGTTGTCGTTGTAAGTTA................................................................................................................................................................................................................................................................................................................ | 22 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................CGGCTTTAGGGCAGCCATGCT.............................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 |
| ............................................................................................AGCTCTCAACCAAATTCCC....................................................................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................AGAGGCTCTCAACCAAATT.......................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group6:12691766-12692107 - | dps_3187 | C------------------------------------------AATTGAAATGGCAGCCATGGCAGCA------GCA---ACAGT------------------CAAT------AACC------ACAAAGGGGTAAGTC---------------------------------------TGACGCGT----CTGCTTTAACTTGGCCTTTAAGC--CG--CTCCGAGGGTTGGTTTACGGGACATT----------GCTT-A----------TG--------ACACACGCGGTCGCCAGACAAATACGCGGCCAG--------------------------------------------------------------------------------------TGTGGCA------------CATGCATTAGTGTGTG-----------------------------TGTATGTATTTGC------------AAACATGCATATTTTATTGCA---------------------------------------------TTGCTCAGGGAGATGTTTTTA-------------------------ATTAACGGGTGCGTAAATCGATCGAA-----AGG-------------GTGATCCA-----AACTTCAA----TTTCTTTCCGCAGCCCAAATCCCGTCGCAACGAGCGCAACATATTGACCTTCTACGAGAAGAT--- |
| droPer2 | scaffold_33:713281-713623 - | C------------------------------------------AATTGAAATGGCAGCCATGGCAGCA------GCA---ACAGT------------------CAAT------AGCC------ACAAAGGGGTAAGTC---------------------------------------TGACGCGT----CTGCTTTAACTTGGCCTTTAAGC--CG--CTCCGAGGGTTGGTTTACGGGACATT---------TGCTT-A----------TG--------ACACACGCGGTCGCCAGACAAATACGCGGCCAG--------------------------------------------------------------------------------------TGTGGCA------------CATGCATTTGTGTGTG-----------------------------TGTATGTATTTGC------------AAACATGCATATTTTATTGCA---------------------------------------------TTGCTCAGGGAGATGTTTTTA-------------------------ATTAACGGGTGCGTAAATCGATCGAA-----AGG-------------GTGATCCA-----AACTTCAA----TTTCTTTCCGCAGCCCAAATCCCGTCGCAACGAGCGCAACATATTGACCTTCTACGAGAAGAT--- | |
| droWil2 | scf2_1100000004768:306445-306502 - | T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCAACAGGCAAAATCACGTCGAAATGAACGTAATATTTTAACATTCTATGAAAAAAT--- | |
| droVir3 | scaffold_13049:9525875-9526066 - | CAT-------ATGGCG-AGCAGTT---------------------------CAGCAGCCATGGACACAGCGACGGCAACAACTACAACAAAAAG---AGGCAAC------------AACAAGAGCAAAGGGGTAAGTGA----GAA--TGT----C----TCG--------------TGC----T----GTA--TTAACTTGGCCTTTAAGC--TGT---GCGAGGGTTGGTTTGTTGTGCGT--T-----TTTGCTT-A----------TGG-CACCATACACGCG-----------------C------------------ATTCGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGACAAACA--- | |
| droMoj3 | scaffold_6680:11236774-11236997 + | CAT-------TTGGCG-AGCAACTTA-------------------------ACACAGCCATGGACACA------GCAGCAGCAACCACGAAAAAGGGAAATAACAACA------ACAGCAAGAGCAAAGGGGTAAGTCA----AAG--TGA----T----TCGTGTTGCTCG-----TGT----T----GTG--TTAACTTGGCCTTTAGCC--AGC---ATGAGGGTTGATTTGTTATGCGT--TT------TGCTT-A----------TGGGCATCATACACACG-----------------C------------------ATTCGTAAGGCAAGCAATTAGTCAC---------------------------------------------------AATGGCG------------TA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droGri2 | scaffold_15110:13059992-13060255 + | CAACTGCTT-TCGGCGCATCAGCTTAGCAGCCATGGACACTGCAGCGGCTGAGGCAGCGACAGCGGCA------GCATCAGCAACCACAAAAAA---AGG---CAAC------AACA------AGAAAGGGGTAAGTTG----AAA--TTT----C----TCA-------C------ACAATTTT----CTG--TTAACTTGGCCTTTAAGC--TGG---ACGGGGGTTGATTTGTTATGCGT--T-----GTTGCCT-A----------TG--------ACGCACAC-------AGACACACACGCACTCGCAC--------ATTCGTAAGACAAACACTT--------------------------------------------------------------------------------------------------------AAGCGCAGTGGCATATCCTGT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATAT--- | |
| droAna3 | scaffold_13337:10166501-10166565 - | AAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACTCTCCCCAGGCCAAATCGCGGCGGAATGAGCGCAATATCCTTACGTTCTACGAAAAGAT--- | |
| droBip1 | scf7180000396569:1193643-1193799 + | C------------------------------------------T---------------ATAGCAGCA------TTGCCAACGGCGACACTGCG---GCGCAATAAAA------ACAACAGCAACAAGGGGGTAAGTAA----GAACTGGA----A----GCC--------------TGTAA--------TG--TTAACTTGGCCCTTAAGC--GATGCGAACGGGGTGGATTTTAC-AAGTTGAGACACTT-----TGGGCCG--------------------------TCGCCAGGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302486:1362823-1363076 + | C------------------------------------------AATTGAAATGGCAGCCATAGCAGCA------TTACCAACGGCGACGCTGCG---GCGCAACAAT------AACA------ACAAAGGGGTAAGT---CC-CAAGTGGA----A----GCC--------------AGATA--------TG--TTAACTTGGCCTTTAAGC--GGTCCTAGGAGGGTGGATTTTAT-AGCTTG--------------------------TGG-----------------CCAACAGACAAACACGCGCGTCATCCAAAGACG----------------CACAGGAACAATTACGTAATTGTGTTTGTACATACACTATAC------------------------A----TATACATGCATACA-----------------------------TATGTATATATGTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTTT--- | |
| droFic1 | scf7180000453929:481700-482175 - | C------------------------------------------TATCTAAATGGCAGCTATTGCGGCA------TTACCAACGGCGACGCTGCG---CCGCAACAAT------AACA------ACAAAGGGGTAAGTTGACC-CAAGGGGA----A----GCC--------------AGATA--------TG--TTAACTTGGCCTTTAAGC--GGCCCGCTGAGGGTGGATTTTAC-AACTTACCT----------CAAGTGAGCA-CGCGG-----------------CCGTCAGACACGT------------TGG-------------------------------------------------------------AAATGTATTGTA---TGTACACACTTCCATACATA----CAAACA------CGTC-------------------------------------CATGTATAGCGTATTAAATATGTATTTACCCTTGAGCCTTGAAGGTCAAGGATTTCTATATACAGTTCTTTGGTATGATCCCTTT-GTCAAAAGGATGTTTTTATACCCTCTCAGACTTTTTGAGATACTATAATAG-ATAGTAAAAT-------AAAATATGAATTTTTAATTGTTTAATTTGTAATCATATTTTACTCTTATTTTATCGTAGATCAAATCTCGTCGCAATGAACGTAACATCTTAACTTTCTACGAGAAGAT--- | |
| droEle1 | scf7180000491193:2004954-2005215 + | C------------------------------------------TATTGAAATGGCAGCTATAGCGGCA------TTACCAACGGCGACGCTGCG---GCGAAACAAT------AACA------ACAAAGGGGTAAGTATTCA-GAAGTGGA----A----GCC--------------AGATATGTTTGTATG--TTAACTTGGCCTTAAAGC--GGTCCGCTGAGGGTGGATTTTAC-AACTGGCCT----------CAAGTGTACA-CGCGG-----------------CCGTCAGACAAGCACGCGCCTAAGCTGGAAGCA---------------------------------------------------------------------------CA----TA------------TGTACATACATATGTATTGTATATACTTATGTACATTTGTAAATACATA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATTT--- | |
| droRho1 | scf7180000758273:960-1119 + | TAC-------TTA--------------------------------TTC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA------------AAACGCATTTACTTTATCGCT---------------------------------------------TTTTTCACG---------------------------------TTAATTTAATAAAATTGTAAAAC-------ATTACATG-------------CTAATTTAGACTGATCTTTTATGCTCTTTTTTCCTTAGGTCAAATCCCGTCGCAATGAGCGTAACATTCTAACTTTTTACGAGAAAAT--- | |
| droBia1 | scf7180000302193:4695024-4695287 + | GAATTTAGTTAAAACT-A------------------------CTACCGAAATGGCAGCTATTGCGGCA------TTGCCAACAGCCCCGTTGCG---GCGCAACAAC------ACCA------GCAAAGGGGTAAGTAGACC-CG-GGGGA----A----GCC--------------AGATAGGT----TGG--TTAACTTGGCCTTTAAGC--GGCCCGCCGAGGGTGGATTTCAC-AACATGCCT----------CGAGTGGACA-CGCGG-----------------CCGTCAGACAAGCACGCGCCTAAACTGC-------------------------------------------------------------A-------------------TGTACTACATACATACGCAGATACT-----------------------------------------TATGTATCTAT------------ATACA-GCAC-TTTTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT--- | |
| droTak1 | scf7180000415809:1105196-1105459 + | C------------------------------------------TATTGAAATGGCAGCTATTGCGGCA------TTACCAACAACTACGTTGCG---CCGCAACAAC------AACA------ACAAAGGGGTAAGTTGTCT-CAGGGATGTACAAATGTATG-----TACATATGTATGTAGGT----TTG--TTAACTTGGCCTTTAAGC--GGTCCGCTGAGGGTGGTTTTCAC-AACTTGCCT----------CAAGTGGACA-CGCGG-----------------CCGTCAGACAAGCACGCGTCTAAACTGG-------------------------------------------------------------AAATATAC----ATATTTACAAATGCA------------CATACATATGTATGT-----------------------------ATGTA------------------------------GGCTTTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT--- | |
| droEug1 | scf7180000409711:4675776-4676026 + | C------------------------------------------TATTGAAATGGCAGCTATTGCGGCA------TTACCAACAACTACGTTGCG---TCGCAACAACAACAATAACAGCAACAACAAAGGGGTGAGTAGTCCAGAAGGAGA----A----GCC--------------AGATATGT----TTG--TTAACTTGGCCTTTAAGC--GGTCCGCTGAGGGTGGATTTTAC-AACTTGCCT----------CAAGTGGGCAGCGCGA-----------------GCGTCAGACAAGCACGCGC-TAAACTGT-------------------------------------------------------------AA------------------------A------------TATACATTTGTATGTA-----------------CATATGTATGTACGTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATATCTA | |
| dm3 | chr3L:21339622-21339862 - | dme_312 | C------------------------------------------TATTGAAATGGCTGCTATTGCCGCC------CTGCCAACAACAACGTTGCG---GCGCAACAACA------ACAACAACACCAAAGGGGTAAGTAGTCG-CAAAGGGA--CAAG-GGACC--------------AGATATGT----CTG--TTAACTTGGCCCTTAAGC--GACCCGCTGAGGGTGGATTTTAT-AACGTGTCT----------CGAGTGGACA-CGCGG-----------------CAGTTACACTAA-AC------------------ACG--------------------------------------------------------------------TGTACACAAGCA------------TATGCATACATATGTA-------------------------------------CATGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATACCGCACACAT--- |
| droSim2 | 3l:20874203-20874428 - | C------------------------------------------TATTGAAATGGCTGCAATTGCCGCC------TTGCCAACAACAACGTTGCG---TCGCAACAACA------ACAGCAACACCAAAGGGGTAAGTTGTCG-CAAGGGGA--CAAGGGGACC--------------AGATATGT----CTG--TTAACTTGGCCTTTAAGTGTGGCCCTCTGAGGGTGGATTTTAT-AACGTGCCT----------CGAGTGGACA-CGCGG-----------------CAGTCACACCAATAC------------------ATG--------------------------------------------------------------------TGTACCTAAGCA------------TATACATACGTATGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_11:1276892-1277120 - | CAA-------C--------------------------------TATTGAAATGGCTGCAATTGCCGCC------TTGCCAACAACAACGTTGCG---TCGCAACAACA------ACAGCAACACCAAAGGGGTAAGTTGTCG-CAAGGGGT--AAAGGGGACC--------------AGATATGT----CTG--TTAACTTGGCCTTTAAGTGTGGCCCTCTGAGGGTGGATTTTAT-AACGTGCCT----------CGAGTGGACA-CGCGG-----------------CAGTCACACCAATAC------------------ATG--------------------------------------------------------------------TGTACCAAAGCA------------TATACATACGTATGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | 3L:19187734-19187977 + | C------------------------------------------TACTGAAATGGCTGCTATTGCCGCC------TTGCCAACAACAACGTTGCG---GCGCAGCAACA------ACAGCAACCACAAAGGGGTAAGTTGTCG-CAAGGAAA----A----GCC--------------AGATATGT----CTG--TTAACTTGGCCTTTAAGC--GACCCTTTGAGGGTGGATTTTAT-AACGTGACT----------CGAGTGGACA-CGCGG-----------------CAGTTGCACCAATAC------------------TTC--------------------------------------------------------------------TGTTTGCACGCA------------TATACATACATATGTA-----------------------------TGTATGTATGTAC------------ATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTTTATAT--- | |
| droEre2 | scaffold_4784:21008313-21008557 - | CAA-------ACAACT-A------------------------CTGCTGAAATGGCTGCTATTGCCGCC------TTGCCAACAACAACGTTGCG---GCGCAACAGCA------ACAGCAACACCAAAGGGGTAAGTTGTCG-CAAGGGGA----A----GCC--------------AGATATGT----CTG--CTAACTTGGCCTTTAAG----GCCCTCTGAGGGTGGATTTTAT-AACGTGCCT----------CGAGTGGACA-CGCGG-----------------C-TTCACACCAATGC------------------ATT--------------------------------------------------------------------TGTACACAAGCA------------TATACATATGGATGTA-----------------------------TGTATGTTTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTACGCAT--- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 06/28/2015 at 03:56 PM