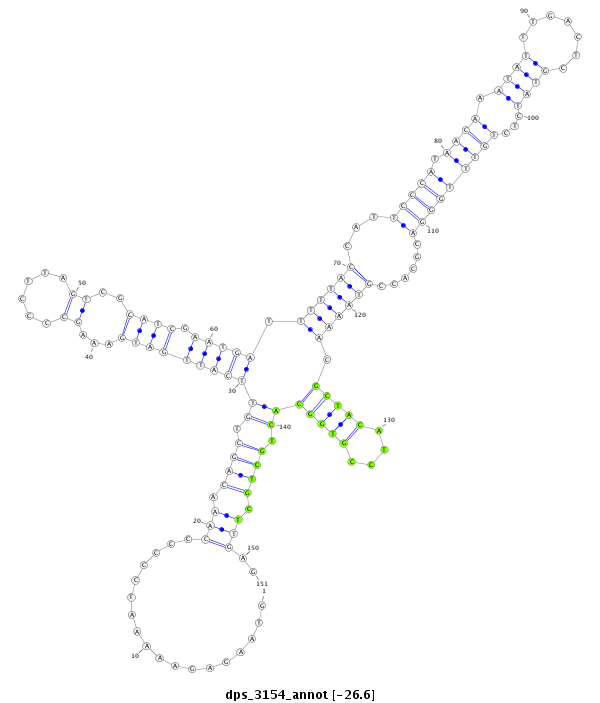
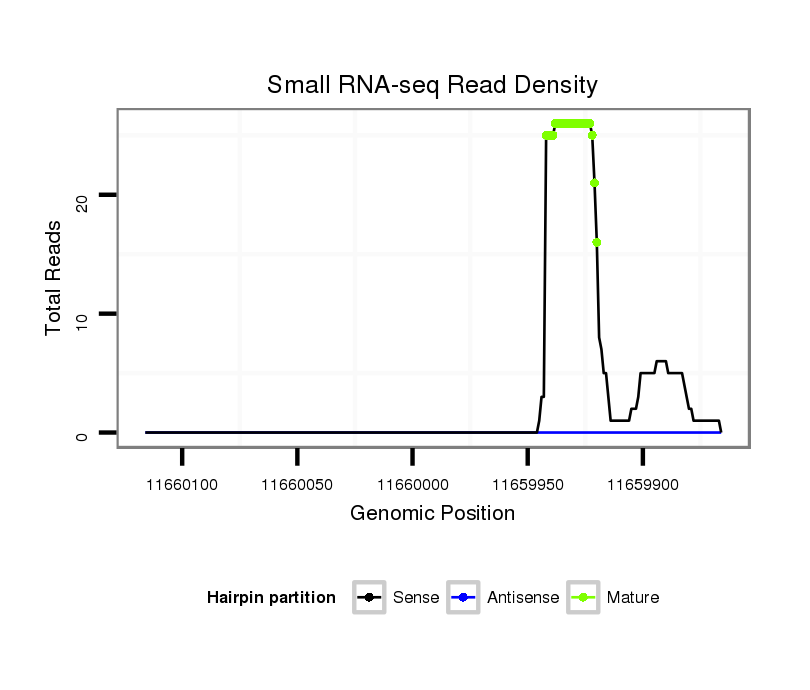
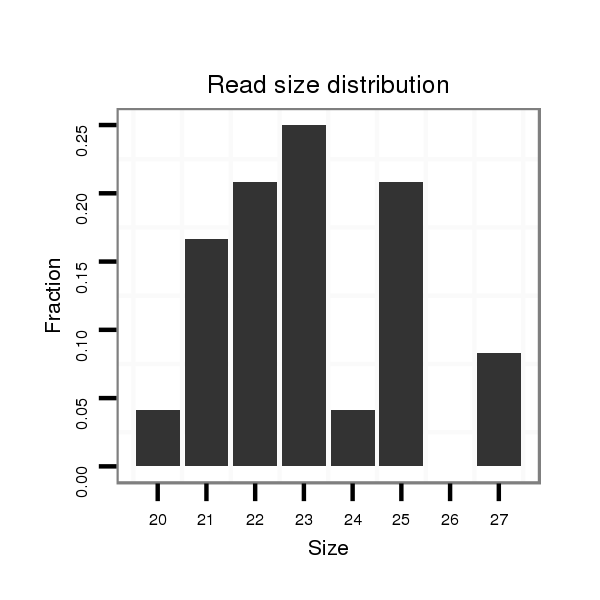
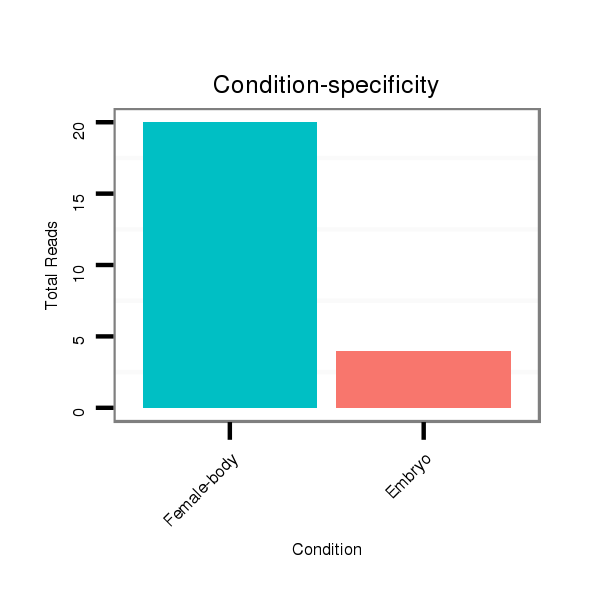
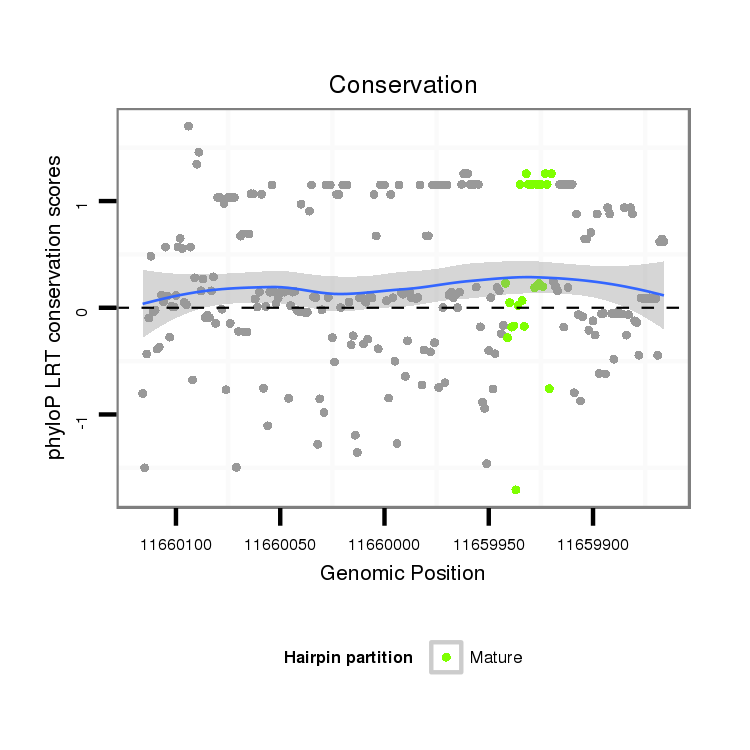
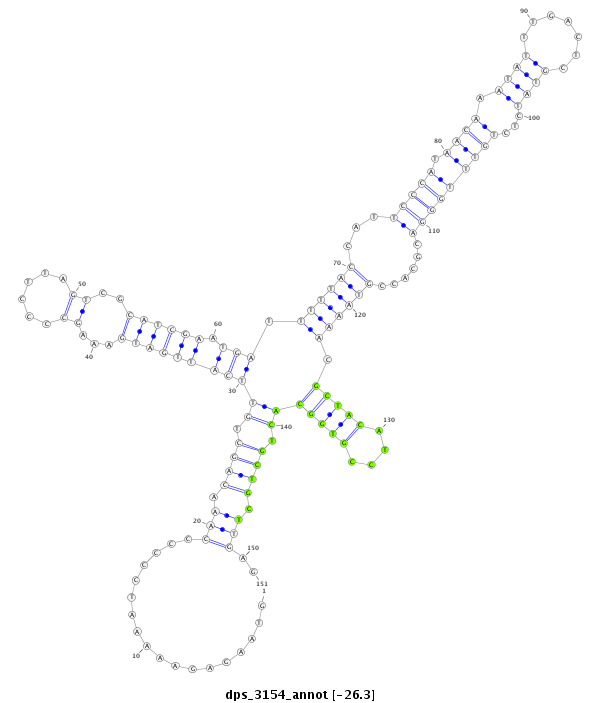
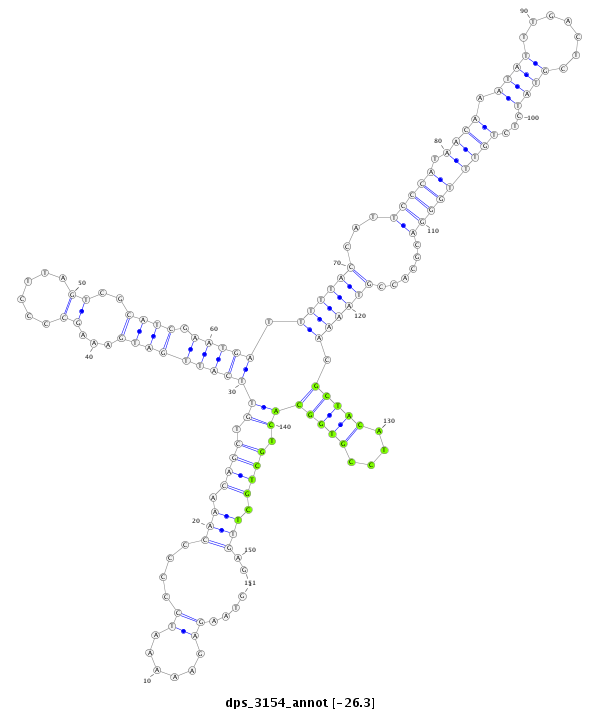
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTGCT...................................................... |
23 |
0 |
1 |
6.00 |
6 |
5 |
1 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTGC....................................................... |
22 |
0 |
1 |
4.00 |
4 |
3 |
1 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTG........................................................ |
21 |
0 |
1 |
4.00 |
4 |
3 |
1 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTGCTTG.................................................... |
25 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
| ............................................................................................................................................................................ACGCTACATCCGTGGCACTGCTGCT...................................................... |
25 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTGCTTGAGG................................................. |
28 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTGCTTGAG.................................................. |
27 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
| ........................................................................................................................................................................................................GGTGACGCCTCAATCAATGCGGGAGCT........................ |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
| ..................................................................................................................................................................................CATCCGTGGCACTGCTGCTTGA................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................................CAATGCGGGAGCTGACCTTATCTG............. |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTGCTT..................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
| ..............................................................................................................................................................................................................................GAGCTGACCTTATCTGAAGAGTGTGAGA. |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ................................................................................................................................................................................................CTGCTTGAGGTGACGCCTT........................................ |
19 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................................AATGCGGGAGCTGACCTTA................. |
19 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
| .......................................................................................................................................................................................................................AATGCGGGAGCTGACCTTAT................ |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ...........................................................................................................................................................................AACGCTACATCCGTGGCACTGCTGC....................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCT......................................................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................AATCAATGCGGGAGCTGACCTTATC............... |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
| ..............................................................................................................................................................................GCTACATCCGTGGCACTGCTGCC...................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
| ............................................................................................................................................................................ACGCTACGTCCGTCGGACTGC.......................................................... |
21 |
3 |
3 |
0.33 |
1 |
0 |
0 |
1 |
0 |
| ...........................................................................................................................................................................AACGCTACGTCCGTTGGACTG........................................................... |
21 |
3 |
3 |
0.33 |
1 |
0 |
0 |
1 |
0 |
| ...............................................................................................................................................................................CTACGTCCGTGATACTGCTGC....................................................... |
21 |
3 |
4 |
0.25 |
1 |
0 |
0 |
1 |
0 |
| .......................................................................ACAGCTGGTCGTTGATGA.................................................................................................................................................................. |
18 |
2 |
9 |
0.11 |
1 |
0 |
0 |
1 |
0 |
| ....................................................................................................................................................................................AGCGTGGCACTGCTGATTG.................................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |