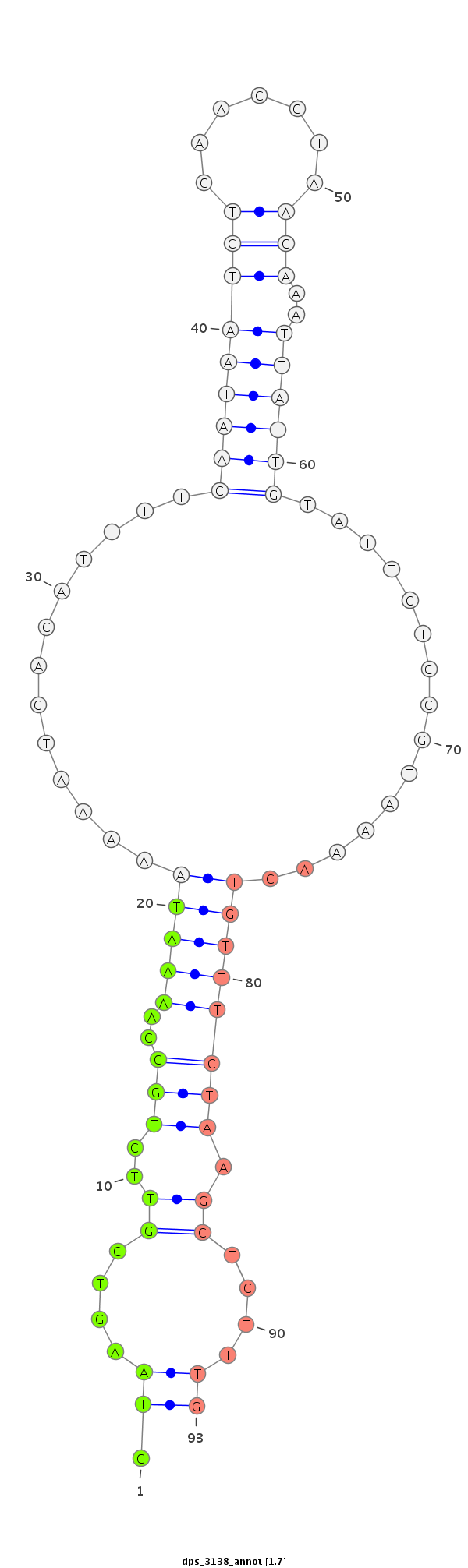

ID:dps_3138 |
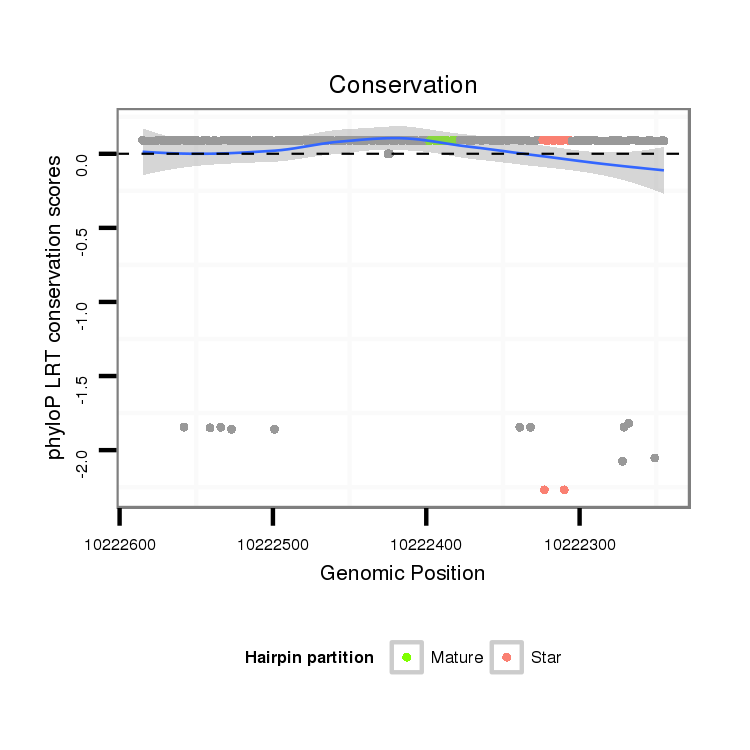
Coordinate:XR_group6:10222295-10222535 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
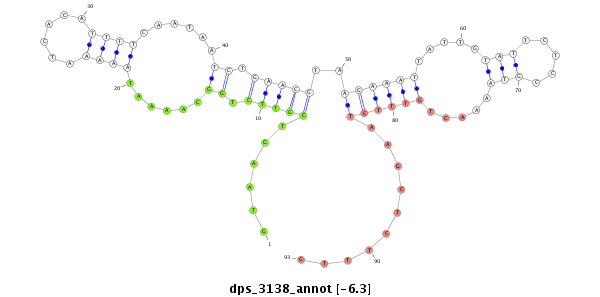
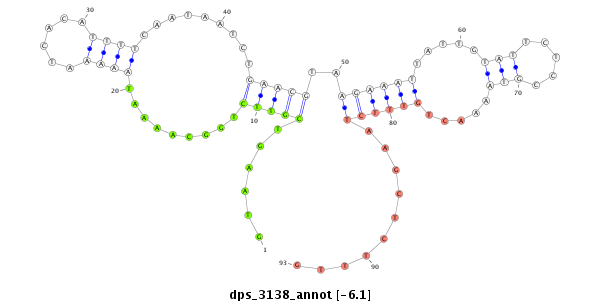
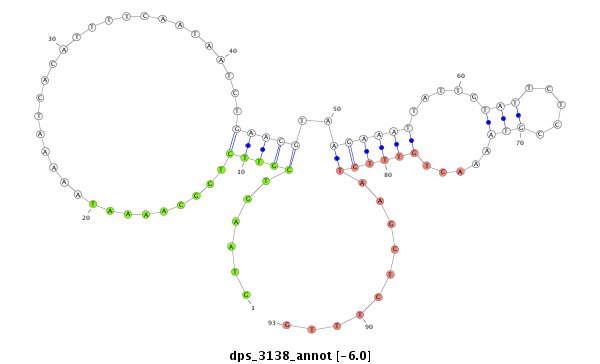
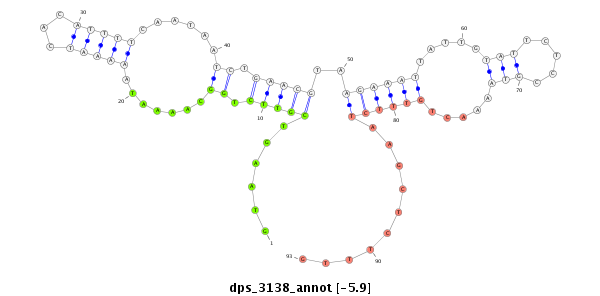
| -6.3 | -6.1 | -6.04 | -5.9 |
 |
 |
 |
 |
exon [dpse_GLEANR_12188:2]; CDS [Dpse\GA23421-cds]; CDS [Dpse\GA23421-cds]; exon [dpse_GLEANR_12188:3]; intron [Dpse\GA23421-in]
No Repeatable elements found
| mature | star |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACGGCGGGGGAAACTTGAAAGTTCAAATCAGAGAAACGGCAAACGGCTGGGTAAAATGCAGTGTTGGAAACCGAGTCAAAGCAAGCCGTGCTGGAAACCGAGTCAAAGGAAGCAGTGCTGGAAACCGAGTCGAAAGAAACTGCTGGAAAACATACCTCTCTCTCTCTTTCATTTCGTGTGCGACGTTGTAAGTCGTTCTGGCAAAATAAAAATCACATTTTCAATAATCTGAACGTAAGAAATTATTGTATTCTCCGTAAAACTGTTTCTAAGCTCTTTGCCGTCTTGCAGGTAATGGCGCCCCGCAGAAGTGATCGAATTTTGGCGGCCAACCCGGCGTC *******************************************************************************************************************************************************************************************.((....((..(((..(((((.............(((((((((.......)))..))))))...............)))))))).))....))************************************************************* |
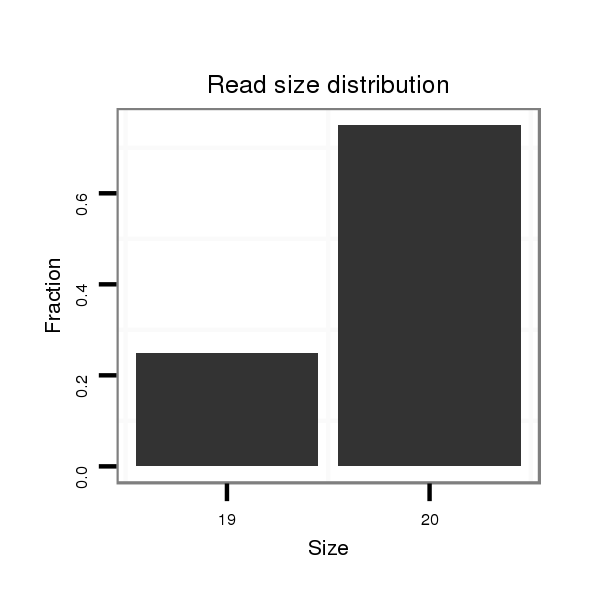
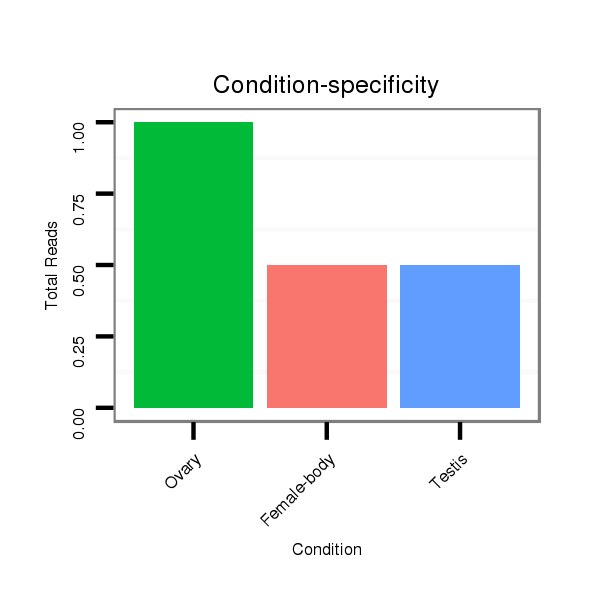
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
SRR902010 ovaries |
M062 head |
V112 male body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................GTAAGTCGTTCTGGCAAAAT...................................................................................................................................... | 20 | 0 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AAGTGATCGAATTTTGGCGG............. | 20 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................AACGGCGAACGGCTCAGTA................................................................................................................................................................................................................................................................................................ | 19 | 3 | 7 | 0.71 | 5 | 0 | 5 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CGTTGTAAGTCGTTCTGGCA.......................................................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................AAGTGATCGAATTTTGGCGGC............ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................CTTGCAGGTAATGGCGCCCC..................................... | 20 | 0 | 4 | 0.50 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................AGTGATCGAATTTTGGCGGC............ | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................TTATCGAATTTTGGCGGCCAACCCGGC... | 27 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................ACTGTTTCTAAGCTCTTTG............................................................. | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................CAAATCAGAGAAACGGCAAAC......................................................................................................................................................................................................................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................TGAACGTAATGGCGCCCCGCAGAAG.............................. | 25 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGAGATGCAGTGTTGGA................................................................................................................................................................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................GTTCAAATCTGCTAAACGG.............................................................................................................................................................................................................................................................................................................. | 19 | 3 | 18 | 0.17 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................CGAATTTTGGAGACCAAC........ | 18 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................AGAGAAACAGCAAAAGGCT..................................................................................................................................................................................................................................................................................................... | 19 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................AGTTCAAATCTGCTAAACG............................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TGCCGCCCCCTTTGAACTTTCAAGTTTAGTCTCTTTGCCGTTTGCCGACCCATTTTACGTCACAACCTTTGGCTCAGTTTCGTTCGGCACGACCTTTGGCTCAGTTTCCTTCGTCACGACCTTTGGCTCAGCTTTCTTTGACGACCTTTTGTATGGAGAGAGAGAGAAAGTAAAGCACACGCTGCAACATTCAGCAAGACCGTTTTATTTTTAGTGTAAAAGTTATTAGACTTGCATTCTTTAATAACATAAGAGGCATTTTGACAAAGATTCGAGAAACGGCAGAACGTCCATTACCGCGGGGCGTCTTCACTAGCTTAAAACCGCCGGTTGGGCCGCAG
*************************************************************.((....((..(((..(((((.............(((((((((.......)))..))))))...............)))))))).))....))******************************************************************************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
GSM444067 head |
SRR902011 testis |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................TTCGCTCAGTTTCGTTCGACACC.......................................................................................................................................................................................................................................................... | 23 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................GAAACGGCAGAACGTCCATTA............................................. | 21 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ......................AGGGTAGTCTCTTTGCCGGTT.......................................................................................................................................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................GGTCAGTTTCCTTCGACACGAT............................................................................................................................................................................................................................. | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................CGTCCATTACCGTGGGGCGTT................................. | 21 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................AGTTTCGTTCGACACGAT........................................................................................................................................................................................................................................................ | 18 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................CATTTTGTCTGGAGAGAGTGA................................................................................................................................................................................ | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................................................................................GTCTAGTACCGCGGGTCGTCT................................ | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group6:10222245-10222585 - | dps_3138 | ACGGCGGGGGAAACTTGAAAGTTCAAATCAGAGAAACGGCAA-ACGGCTGGGTAAAATGCAGTGTTGGAAACCGAGTCAAAGCAAGCCGTGCTGGAAACCGAGTCAAAGGAAGCAGTGCTGGAAACCGAGTCGAAAGAAACTGCTGGAAAACATACCTCTCTCTCTCTTTCATTTCGTGTGCGACGTTGTAAGTCGTTCTGGCAAAATAAAAATCACATTTTCAATAATCTGAACGTAAGAAATTATTGTATTCTCCGTAAAACTGTT-TCTAAGCTCTTTGCCGTCTTGCAGGTAATGGCGCCCCGCAGAAGTGATCGAATTTTGGCGGCCAACCCGGCGTC |
| droPer2 | scaffold_20:155278-155618 - | ACGGCGGGGGAAACTTGAAAGTTCAAAACAGAGAAACGGCAAAACAGCTGGGCAAAATGTAGTGTTGGAAACCGAGTCAAAGCAAGCTGTGCTGGAAACCGAGTCAAAGGAAGCAGTGCTGGAAACCGAGTCGAAAGAAACTGCTGGAAAACATACCTCTC--TCTCTTTCATTTCGTGTGCGACGTTGTAAGTCGTTCTGGCAAAATAAAAATCACATTTTCAATAATCTGAACGTAAGAAATTATCGTATTCCCCGTAAAAGTGTTTTCTAAGCTGTTTGCCGTCTTGCAGGTAATGGCGCCCCGCAGAAGTGCACGTATTTTGGCGGCCAACCAGGCGTC |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 06:17 AM