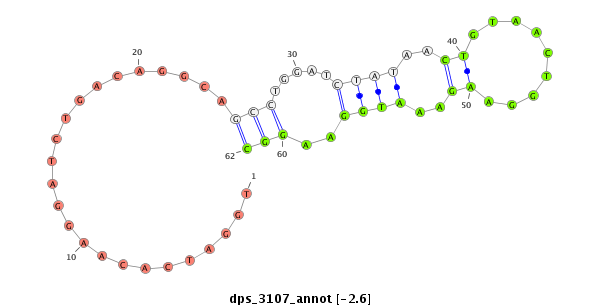
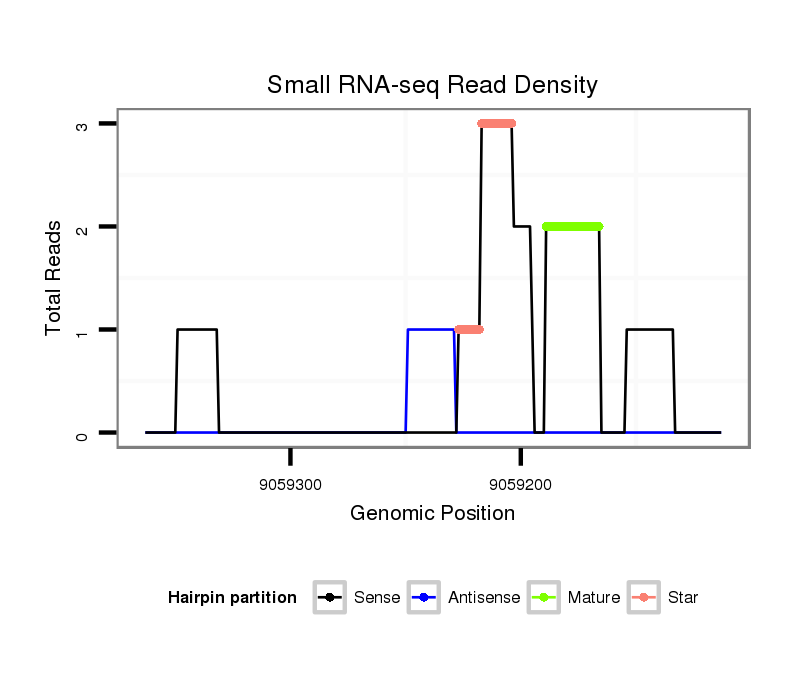
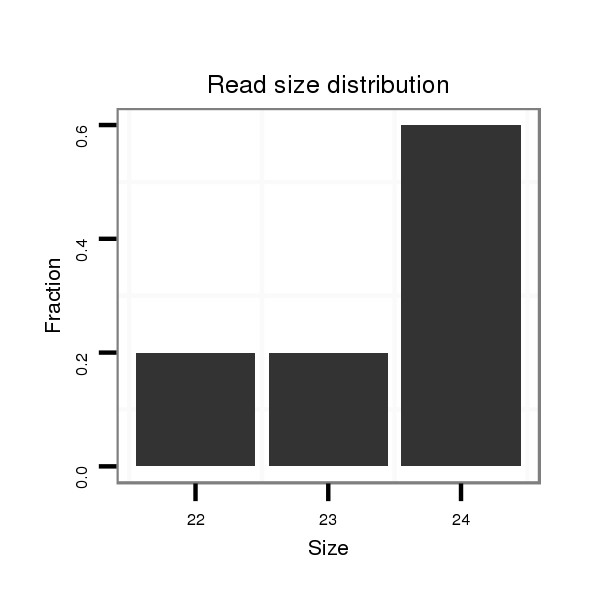
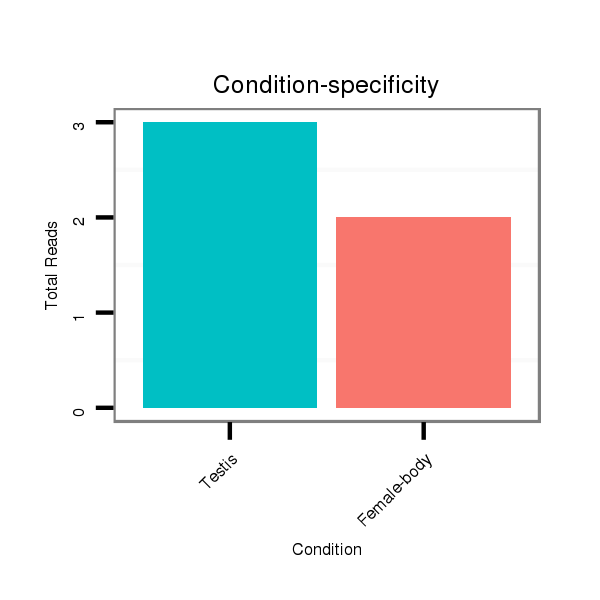
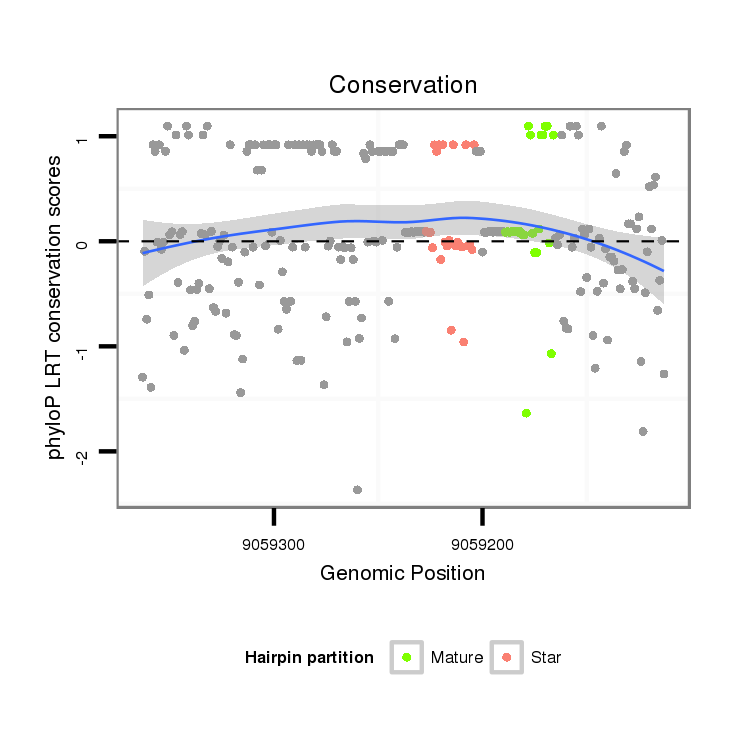
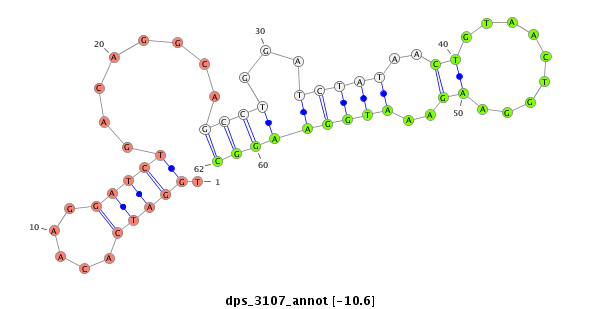
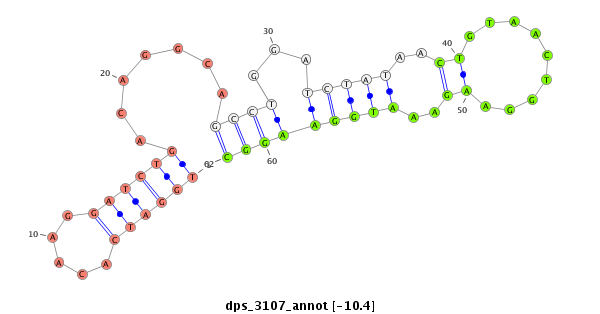
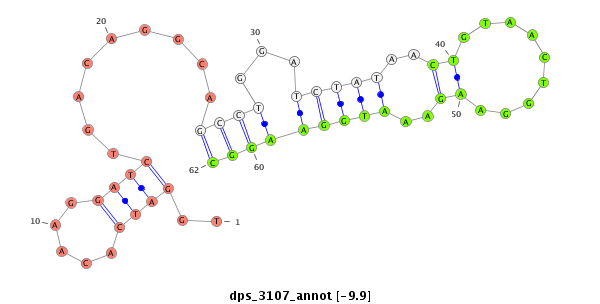
ID:dps_3107 |
Coordinate:XR_group6:9059163-9059313 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -10.6 | -10.4 | -9.9 |
 |
 |
 |
CDS [Dpse\GA23457-cds]; exon [dpse_GLEANR_12285:5]; intron [Dpse\GA23457-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCGTTTGTTTTCTGGTCTGGCTGGATGGAGGTAAGGTAGGTGACCGCTTTGTTCTTCAACTTCAAGCCCAAGCTGTGTGTTCTTTAGGCCTAGCGCAACGACGTGATGTTGAAGGCCCACCCATAAGCGTCTTTAGTGGATCACAAGGATCTGACAGGCAGCCTGGATCTATAACTGTAACTGGAAGAAATGGAAGGCCAGGTGCATCTGTTCAATCCCGTTGTCCTTGCTGTGGTGCCCTTGGAGGGAGC ****************************************************************************************************************************************........................(((.....((((..((.........))..))))..)))***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
GSM444067 head |
V112 male body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................CTGTAACTGGAAGAAATGGAAGGC..................................................... | 24 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................TGGATCACAAGGATCTGACAGGCA........................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GTTCAATCCCGTTGTCCTTGC..................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................GGATCTGACAGGCAGCCTGGAT................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................TCATGTTGAAGCCCCACCCA................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................AGGCAGTGTGGTTCTATAACT........................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............GTCTGGCTGGATGGAGGT........................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................GGATCTGACAGGCAGCCTGGATC.................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................CATCGGTTCAATCCCGGT............................. | 18 | 2 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................GAGCTAAGGAAGGTGACCGGT........................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............GGTCTGGCTGGATGGAGGCCT......................................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................AATGGACGGCCGGGGGCATC........................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........CAGGTCTCGCTGGATGGCGG............................................................................................................................................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................ATGTAGGTAAGGGAAGTGAC............................................................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CAATAGCTTCTTTAGTGGA............................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| ........................AGGGAGGTAAGGTGGGGGAC............................................................................................................................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AATGATCTGACAGGCGGCAT....................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TGACAGGCAGCTTGGTTCA................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................AGGACTCGATGTTGAAGGC....................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
AGCAAACAAAAGACCAGACCGACCTACCTCCATTCCATCCACTGGCGAAACAAGAAGTTGAAGTTCGGGTTCGACACACAAGAAATCCGGATCGCGTTGCTGCACTACAACTTCCGGGTGGGTATTCGCAGAAATCACCTAGTGTTCCTAGACTGTCCGTCGGACCTAGATATTGACATTGACCTTCTTTACCTTCCGGTCCACGTAGACAAGTTAGGGCAACAGGAACGACACCACGGGAACCTCCCTCG
*****************************************************........................(((.....((((..((.........))..))))..)))**************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
SRR902010 ovaries |
|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................CGGGTGGGTATTCGCAGAAAT.................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ........................................................GTTGAAGTTCGGGTAGAACAC.............................................................................................................................................................................. | 21 | 3 | 3 | 1.00 | 3 | 2 | 1 | 0 |
| ..................................................................................................GCTGTAGTACAACTTCCGGGA.................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................GGGCAACAGGAACGATAG................. | 18 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 |
| .........................................................TTGAAGTTCGGGTAGAACAC.............................................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 |
| ........................................................GTTGACGTTCGGGTTC................................................................................................................................................................................... | 16 | 1 | 14 | 0.07 | 1 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................CGGTCCACTTAGGCAATT..................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..................................................................................................................CGGGTAGGTCTTCGCAGGA...................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group6:9059113-9059363 - | dps_3107 | TCGTTTGTTTTCTGGTCTGGCTGGATGGAGGTAAGGTA-GGTGACCG---CTTTGTTCTTCAAC-TTCAAGCCCAAGCTGTGTGTTCTTTAGGCCTAGCGCA--ACGACGTGATG----TTGAAGGCCCACCCATAAGCGTCTTTAGTGGATCACAAGG--ATCTGACAG-----------------------GCAGCCTGGATCTATAACTGTAACTGGAAGAAATGGA------------------------------------------------------AG--GCCAGGTGCATCTGTTCAATCCCGTTGTCCTTGCTGTGGTGCCCTTGGAGGGAGC |
| droPer2 | scaffold_35:667110-667360 + | TCGTTTGTTTTCTGGTCTGGCTGGATGGAGGTAAGGTA-GGTGACCG---CGTTGTTCTTCAAC-TTCAAGCCCAAGCTGTGTGTTCTTTAGGCCTAGCGCA--ACGACGCGATG----TTGAAGGCCCACCCATAAGCGTCTTTAGTGGATCACAAGG--ATCTGACAG-----------------------GCAGCCTGGATCTATAACTGTAACTGGAAGAAATGGA------------------------------------------------------AG--GCCAGGTGCATCTGTTCAATCCCGTTGTCCTTGCTGTGGTGCCCTTGGAGGGAGC | |
| droWil2 | scf2_1100000004515:3267944-3267952 + | G----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAGGAGGT | |
| droAna3 | scaffold_13266:863663-863687 - | TCGTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGGAGCTCCAGGAGGACCT | |
| droKik1 | scf7180000302698:794134-794233 + | ACACG-------TTGCCCTGTTGTGTAAGGATCA------AA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGAGTAATGGA------------------------------------------------------AGCATCAAAGTTTATATGCTTTATGTTTTCACTTTTAACGTTTTTCTTTTGGAGGAGGA | |
| droFic1 | scf7180000453841:421260-421260 + | C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droEle1 | scf7180000490564:1473276-1473507 - | TAATCTGCTGCCTTAGCTTGTTGCCCGAAGGTGATATT-TAGGAAAA---CTTTGTTAT---ACATT-AACTTCTAAATTTATTTTCTTTAGAAAAAGCTGA--GGAGGATTT----TCCTGTGGGTCCGCCCATAA-------------GTCAGAGGAGACTCCAAAAGTTTAGCTCCTTTGGCTTGGCAGGGCAGCCT------------------GGCAGACCTGGA------------------------------------------------------AT--GCCTGGCCGACCTGGTCGACCGGGTCAGCCA------GGAACTCCTGGCGG---- | |
| droRho1 | scf7180000780091:268232-268514 - | TAATCTGCTGCCTTGGCTTAATGCACAGAGGTGATAAT-----AAGCAAGTTATGTTAT---ACATT-AACTTTAAGATGTGTTTTCTTTAGGTCAAGCTGTCGAAGAGGAGGTGTTTCCTGTGGGTCCACCAATAA-------------ATCAGAGGAGTATCCGAGAGGTAAGCTCATTTGCCTCGCCAGGACAGCCT------------------GGTAGACCTGGACAAGGAATACCTGGCCGACCTGGTTCGGGAATACCTGGCCGACCTGGTTCGGGAAT--ACCTGGCCGACCTGGTCGACCAGGTC------------------AACCAGGAAGT | |
| droBia1 | scf7180000302428:7113497-7113721 + | TAGTCTGCTGCCTTGCCCTGATGCAGGAAGGTGCACTTCTAAAAAAA-----CGGTTATTAAAT-TT-AAATCTTAAATATTTTTTCTTTAGCTCAAGCTGACGAGAAGAATGTGCTTCCTGTGGGTCCGCCAGTAA-------------ATCAGAGAAGTCTTCGTGAAGTAGGCTCCTTTGCCAGAGAAGGGAAGCCG------------------GGAAGACCCGGA------------------------------------------------------AG--ACCTGGGTTACCTGGCCGGCGAGGTA------------------------AGGCC | |
| droTak1 | scf7180000416006:26633-26649 - | GCCCT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAAGATGGGGA | |
| droSec2 | scaffold_1:4724184-4724192 - | G----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCGGAGGC | |
| droYak3 | 3L:4509701-4509723 + | GCGGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGTGGTGGAGGAGGCGGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/18/2015 at 06:06 AM