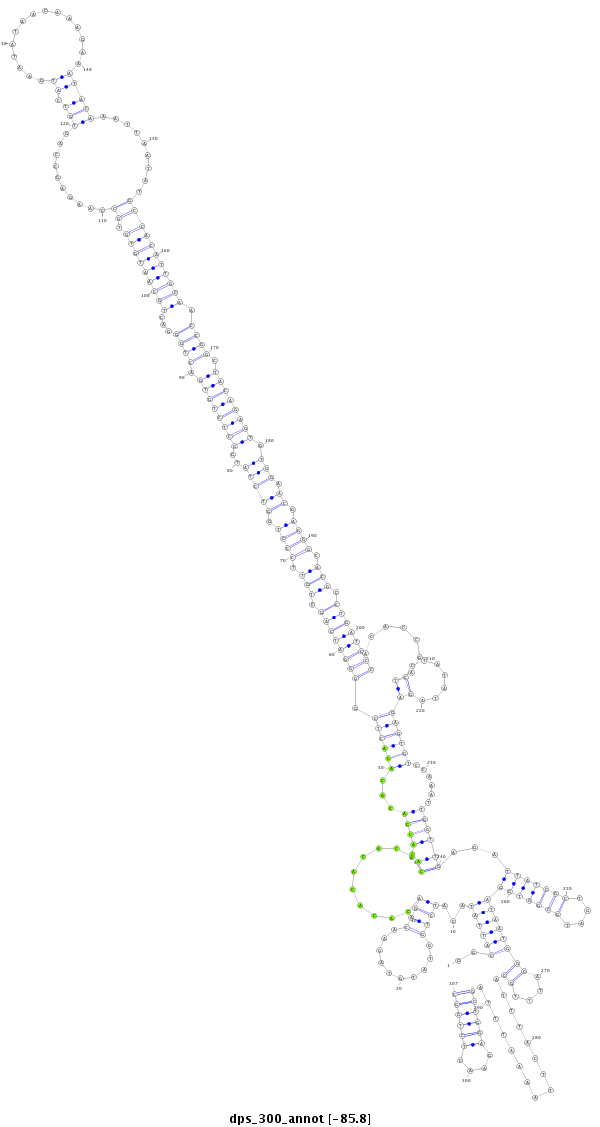
| .......................................................................................CGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
18 |
2 |
1 |
3447.00 |
3447 |
3422 |
25 |
0 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
2 |
1 |
546.00 |
546 |
525 |
21 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
19 |
3 |
15 |
484.13 |
7262 |
7092 |
169 |
1 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
20 |
3 |
4 |
229.00 |
916 |
822 |
94 |
0 |
0 |
0 |
| .....................................................................................AACGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
3 |
2 |
134.00 |
268 |
243 |
25 |
0 |
0 |
0 |
| .......................................................................................CTCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
18 |
1 |
1 |
42.00 |
42 |
41 |
0 |
1 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGCGGTT................................................................................................................................................................................................................................................................................. |
18 |
2 |
1 |
16.00 |
16 |
15 |
1 |
0 |
0 |
0 |
| .......................................................................................CTCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
19 |
2 |
4 |
11.25 |
45 |
32 |
13 |
0 |
0 |
0 |
| ......................................................................................ACCCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
2 |
1 |
8.00 |
8 |
5 |
3 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCTGTTC................................................................................................................................................................................................................................................................................ |
18 |
1 |
1 |
5.00 |
5 |
5 |
0 |
0 |
0 |
0 |
| ......................................................................................ACTCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
1 |
1 |
4.00 |
4 |
3 |
1 |
0 |
0 |
0 |
| .......................................................................................CCCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
18 |
2 |
3 |
4.00 |
12 |
10 |
2 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCGGTTCC............................................................................................................................................................................................................................................................................... |
19 |
2 |
1 |
4.00 |
4 |
2 |
2 |
0 |
0 |
0 |
| .....................................................................................AACTCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
2 |
1 |
3.00 |
3 |
2 |
1 |
0 |
0 |
0 |
| ......................................................................................ACTCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
20 |
2 |
3 |
2.67 |
8 |
7 |
1 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCTGTTCG............................................................................................................................................................................................................................................................................... |
19 |
2 |
2 |
2.00 |
4 |
4 |
0 |
0 |
0 |
0 |
| ......................................................................................ATGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
15 |
1.67 |
25 |
17 |
8 |
0 |
0 |
0 |
| ......................................................................................ACGTGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
8 |
1.63 |
13 |
8 |
5 |
0 |
0 |
0 |
| ........................................................................................GCGGGGATCAGCGGTTCC............................................................................................................................................................................................................................................................................... |
18 |
2 |
2 |
1.50 |
3 |
1 |
2 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCGGTTCGC.............................................................................................................................................................................................................................................................................. |
20 |
3 |
2 |
1.50 |
3 |
3 |
0 |
0 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGTGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
9 |
1.22 |
11 |
9 |
2 |
0 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGCTGTTCG............................................................................................................................................................................................................................................................................... |
20 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................ACTCGGGGATCAGCGGTT................................................................................................................................................................................................................................................................................. |
18 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................CACCCGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
22 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................ACTCGCGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ................CTTGTGGTTAACAGCAGCGGG.................................................................................................................................................................................................................................................................................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| .....................................................................................AACGCGGGGATCAGCTGTTCG............................................................................................................................................................................................................................................................................... |
21 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................CACGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
18 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGCGGTTCC............................................................................................................................................................................................................................................................................... |
20 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .............................................................................................................................................................................................................................................................................................................................................................TGGATTGTAATCATGCTCGTCG...... |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCCGTTC................................................................................................................................................................................................................................................................................ |
18 |
2 |
3 |
1.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| .....................................................................................AACCCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
3 |
3 |
1.00 |
3 |
2 |
1 |
0 |
0 |
0 |
| .......................................................................................CCCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.75 |
15 |
11 |
4 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCGGTTT................................................................................................................................................................................................................................................................................ |
18 |
3 |
20 |
0.55 |
11 |
11 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................................................................................................................................TTGCGGTGGATGATGGCATTGGC................................................................... |
23 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................ACGCGGGGATCACCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
6 |
0.50 |
3 |
2 |
1 |
0 |
0 |
0 |
| .....................................................................................CTCGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................AACGCGGGGATCAGCGGTT................................................................................................................................................................................................................................................................................. |
19 |
3 |
9 |
0.44 |
4 |
4 |
0 |
0 |
0 |
0 |
| ......................................................................................ACACGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
20 |
3 |
7 |
0.43 |
3 |
2 |
1 |
0 |
0 |
0 |
| ......................................................................................CCGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
14 |
0.43 |
6 |
6 |
0 |
0 |
0 |
0 |
| ......................................................................................ACCCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
20 |
3 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
0 |
| ......................................................................................TCGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
10 |
0.40 |
4 |
4 |
0 |
0 |
0 |
0 |
| .....................................................................................CGCGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................AACTCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
21 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................AACACGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................CTCGGGGATCAGCGGTTCA............................................................................................................................................................................................................................................................................... |
19 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................CACGCGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
21 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................TACGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................CGGGGATCAGCGGTTTCC.............................................................................................................................................................................................................................................................................. |
18 |
2 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................CATGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
20 |
3 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................GCGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
17 |
0.24 |
4 |
4 |
0 |
0 |
0 |
0 |
| ........................................................................................GCGGGGATCAGCGGTTCGC.............................................................................................................................................................................................................................................................................. |
19 |
3 |
20 |
0.20 |
4 |
3 |
1 |
0 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGCGGTTT................................................................................................................................................................................................................................................................................ |
19 |
3 |
10 |
0.20 |
2 |
2 |
0 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCAGTTC................................................................................................................................................................................................................................................................................ |
18 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................ACGCGCGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
11 |
0.18 |
2 |
2 |
0 |
0 |
0 |
0 |
| ....................................................................................ACAGTCGGGGATGAGCTGTA................................................................................................................................................................................................................................................................................. |
20 |
3 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................ACGCGGGGATCAGCGGTCC................................................................................................................................................................................................................................................................................ |
19 |
3 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................................................................................................................................................GCTGGAGAAATCTGGCGC................................. |
18 |
2 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................ACCGGGGATCAGCGGTTCC............................................................................................................................................................................................................................................................................... |
19 |
3 |
9 |
0.11 |
1 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................CACGGGGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
| .......................................................................................ACCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
18 |
3 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
0 |
| .......................................................................................CGCGGGGATCAGCGGTTCA............................................................................................................................................................................................................................................................................... |
19 |
3 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................GCGGGGATCAGCTGTTCG............................................................................................................................................................................................................................................................................... |
18 |
2 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................AAGCGGGGATCAGCGGTTC................................................................................................................................................................................................................................................................................ |
19 |
3 |
15 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................ATGGTCTATGGCTCTGCGCC........................................................................................................................................................................................................................................................... |
20 |
3 |
18 |
0.06 |
1 |
0 |
0 |
0 |
0 |
1 |
| .......................................................................................CTCGGGGATCAGCGGGTCG............................................................................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................AGTCGGGGATGAGCTGTTG................................................................................................................................................................................................................................................................................ |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................AAAGTCGGGGATTAGCTGT.................................................................................................................................................................................................................................................................................. |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................CACACACACACACACCACACACA.................................................................................................................................................................................................................................................................................................. |
23 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................CTCGGTGATCAGCGGTTCG............................................................................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................ATGCGGGGATCAGCGGTT................................................................................................................................................................................................................................................................................. |
18 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................CGGGGATCAGCGGTTCGTT............................................................................................................................................................................................................................................................................. |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................................CTCGGGGATCAGTGGTTCG............................................................................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |