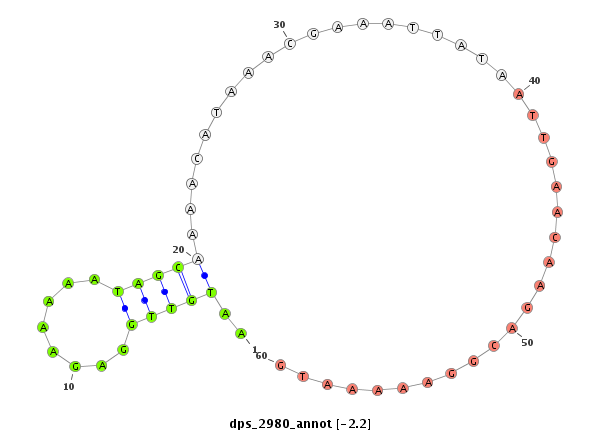
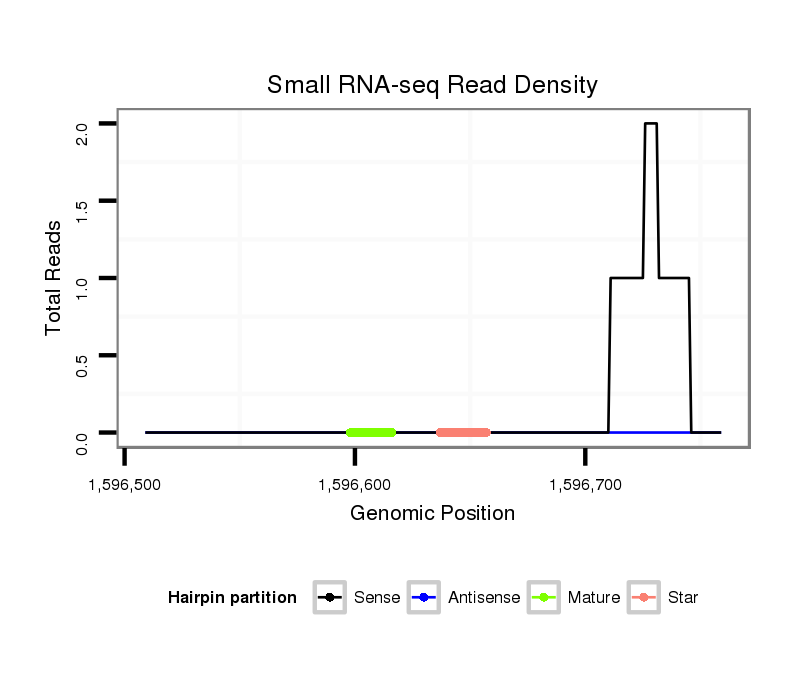

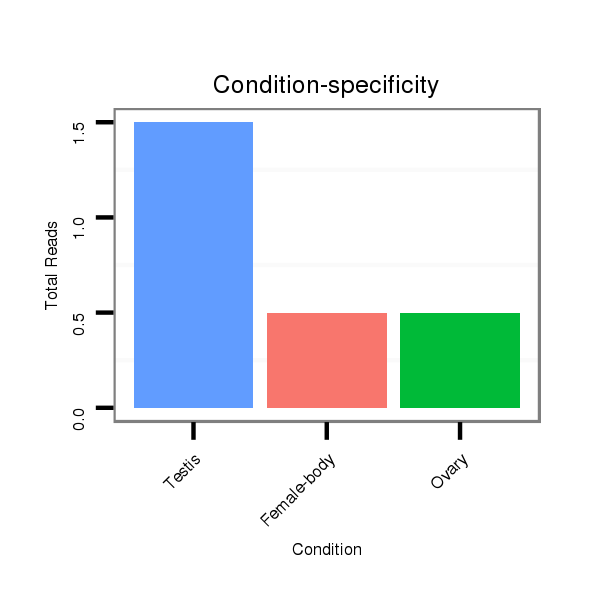
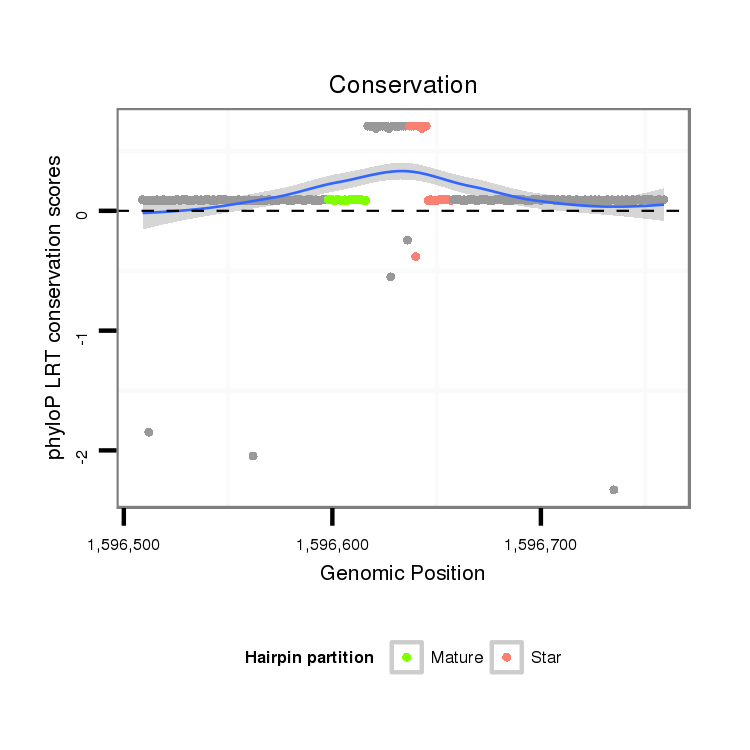
ID:dps_2980 |
Coordinate:XR_group6:1596559-1596709 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -1.2 | -1.2 |
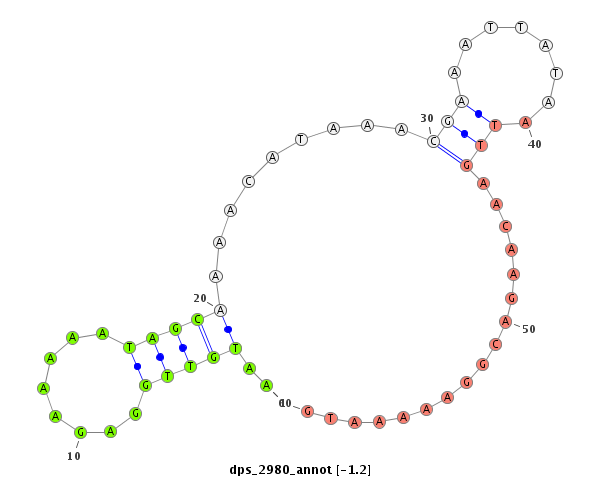 |
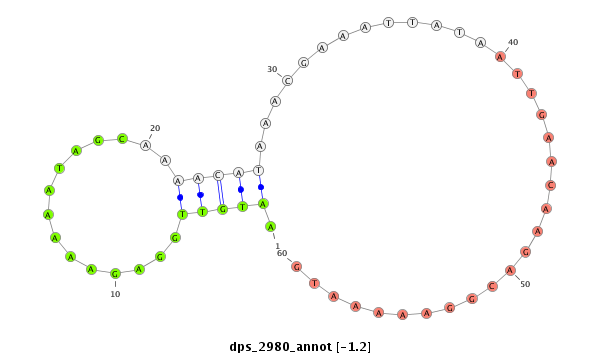 |
exon [dpse_GLEANR_12858:3]; CDS [Dpse\GA23675-cds]; intron [Dpse\GA23675-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------#######################################----------- TCGCCGCAGCACGGCCATGAATCGGAAGAGAAACTCTTTTTGGGGGACGGCAGAGGCAGCAACGGGAAAAGCCATTTGCATACACGAAAAATGTTGGAGAAAAATAGCAAAACATAAACGAAATTATAATTGAACAAGACGGAAAAATGAATTCTCATTTGCTAAAGCGATTAAGGCGATTAAAGCGATTACATTTTACAGATGTACGAGGGACGAGAGACGGAGAGGAGAGACGAGTAGTTTTTGGTAGA *****************************************************************************************..(((((........)))))........................................****************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
V112 male body |
SRR902010 ovaries |
M022 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............CCATGAGTCGGAAGCCAAAC......................................................................................................................................................................................................................... | 20 | 3 | 4 | 2.00 | 8 | 8 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TGTACGAGGGACGAGAGACGG............................ | 21 | 0 | 2 | 1.50 | 3 | 0 | 3 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................AGACGGAGAGGAGAGACGAG.............. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................CTCTTATTGGGTGACGGC........................................................................................................................................................................................................ | 18 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...............CATGAGTCGGAAGCCAAAC......................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.90 | 18 | 14 | 0 | 2 | 0 | 2 |
| .............................................................................................................................................................TTTGCTAAAGCGATTAGGGC.......................................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................AATGTTGGAGAAAAATAGC............................................................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................ATTGAACAAGACGGAAAAATG...................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................AAATTATAATTGAACAAGACGG............................................................................................................. | 22 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................TTATAATTGAACAAGACGGAA........................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TTTTGCAGATGTACGAGGGA...................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................TTTACAGATGTACGAGGGACG.................................... | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................ATTGTAATTGAACAAGACGGA............................................................................................................ | 21 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................TGCATACACGAAAAATGTTGG.......................................................................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................TTGGGCGACGGTAGAGGCTGC............................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............CATGAGTCGGAAGCCAAACT........................................................................................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................ACTCTTATGGGGTGACGGC........................................................................................................................................................................................................ | 19 | 3 | 16 | 0.19 | 3 | 3 | 0 | 0 | 0 | 0 |
| .................................CTCTTATGGGGGGACGGC........................................................................................................................................................................................................ | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............CCATGAGTCGGAAGCCAAA.......................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................ACGAAAAATGGTGGAGGAAAT................................................................................................................................................... | 21 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TAATCGGACAAGACAGAAAAA........................................................................................................ | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................TACAAATGCACCAGGGACG.................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................TGGGCGACGGTAGAGGCTGC............................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
AGCGGCGTCGTGCCGGTACTTAGCCTTCTCTTTGAGAAAAACCCCCTGCCGTCTCCGTCGTTGCCCTTTTCGGTAAACGTATGTGCTTTTTACAACCTCTTTTTATCGTTTTGTATTTGCTTTAATATTAACTTGTTCTGCCTTTTTACTTAAGAGTAAACGATTTCGCTAATTCCGCTAATTTCGCTAATGTAAAATGTCTACATGCTCCCTGCTCTCTGCCTCTCCTCTCTGCTCATCAAAAACCATCT
******************************************************************************************************..(((((........)))))........................................***************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|
| ..............GGTACTCAGCCTTCGGTTTG......................................................................................................................................................................................................................... | 20 | 3 | 4 | 2.50 | 10 | 10 | 0 | 0 |
| ....................................AAAAAGCCGCTGCCGTCTCC................................................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .............CGGTACTCAGCCTTCGGTTTG......................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ..................................................................................................................ATTTGCTTTAATATTAACTT..................................................................................................................... | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| .....................................AAAAACCGCTGCCGTCTCC................................................................................................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ...GACGTCGTGCCGGTCCTTAG.................................................................................................................................................................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................CTCCCTTCTCTCTGACACTCC....................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 |
| ....................................................................................................................TTGCTATAATATTAACTGTTT.................................................................................................................. | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group6:1596509-1596759 + | dps_2980 | TCGCCGCAGCACGGCCATGAATCGGAAGAGAAACTCTTTTTGGGGGACGGCAGAGGCAGCAACGGGAAAAGCCATTTGCATACACGAAAAATGTTGGAGAAAAATAGCAAAACATAAACGAAATTATAATTGAACAAGACGGAAAAATGAATTCTCATTTGCTAAAGCGATTAAGGCGATTAAAGCGATTACATTTTACAGATGTACGAGGGACGA--------------------GAGACGGAGAGGAGAGACGAGTAGTTTTTGGTAGA |
| droPer2 | scaffold_12:2286977-2287247 + | TCGTCGCAGCACGGCCATGAATCGGAAGAGAAACTCTTTTTGGGGGACGGCAGCGGCAGCAACGGGAAAAGCCATTTGCATACACGAAAAATGTTGGAGAAAAATAGCAAAACATAAACGAAATTATAATTGAACAAGACGGAAAAATGAATTCTCATTTGCTAAAGCGATTAAGGCGATTAAAGCGATTACATTTTACAGATGTACGAGGGACGAGAGATGGCGACGAGAGACGAGAGACGGAGACGAGAGACGAGTAGTTTTTGGTAGA | |
| droEug1 | scf7180000409215:73776-73804 + | ------------------------------------------------------------------------------------------------------------AAAACATAAACCAAATTATTATTTAACAA-------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
Generated: 05/18/2015 at 05:56 AM