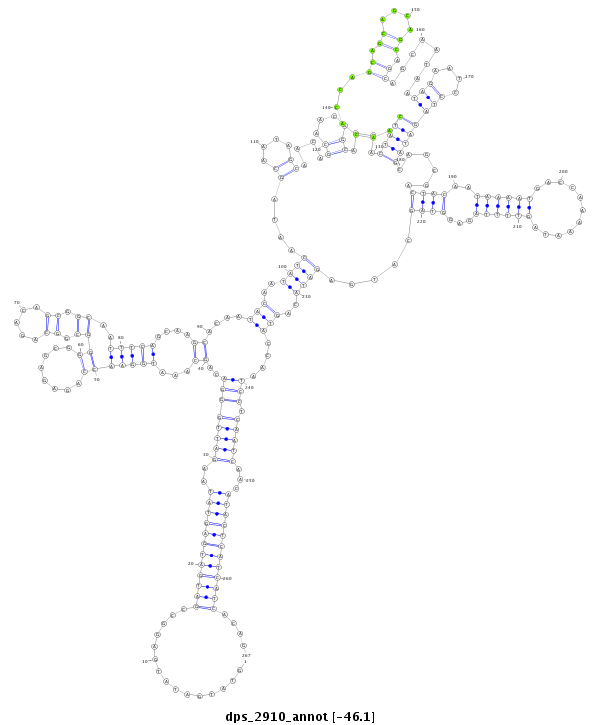
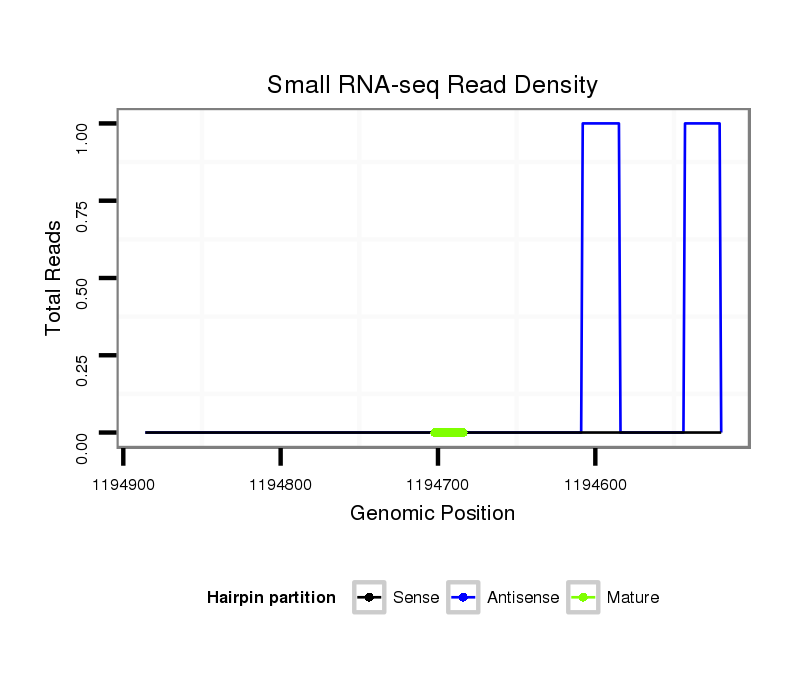
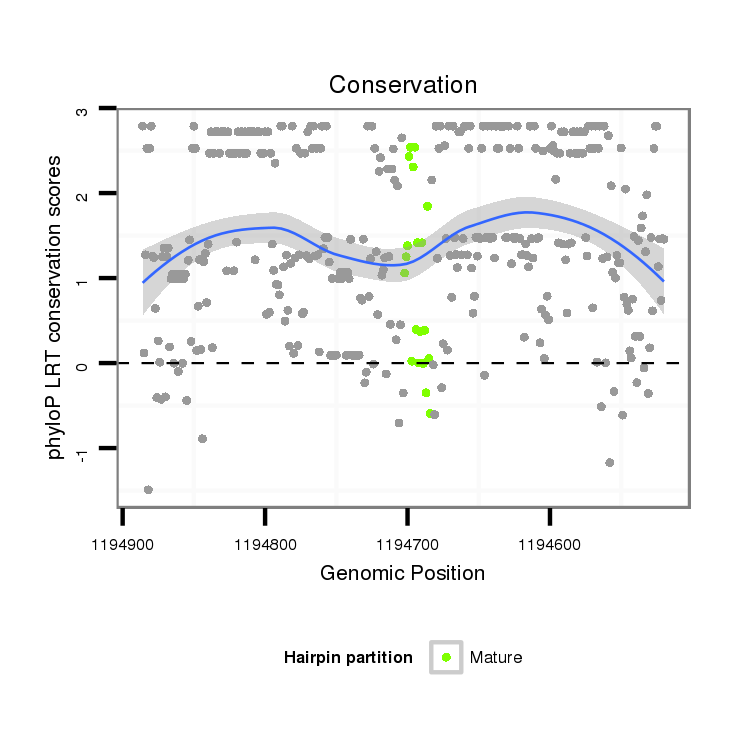
ID:dps_2910 |
Coordinate:XR_group3a:1194570-1194836 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -46.1 | -45.9 | -45.9 |
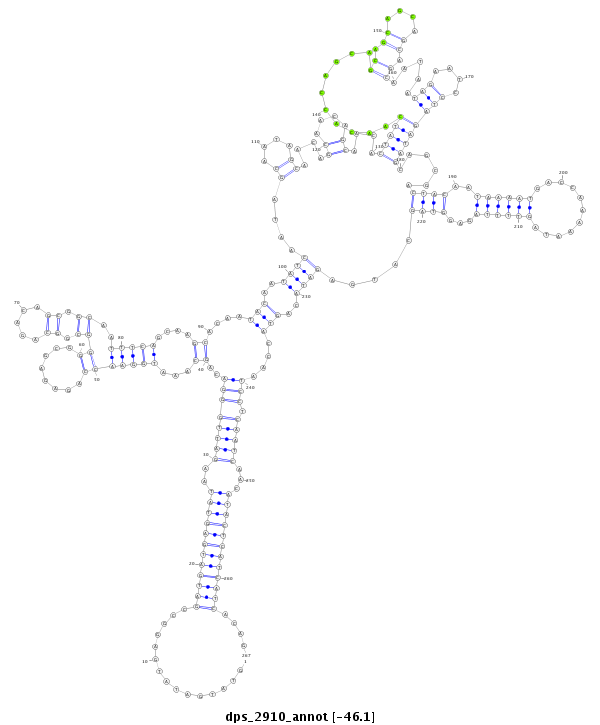 |
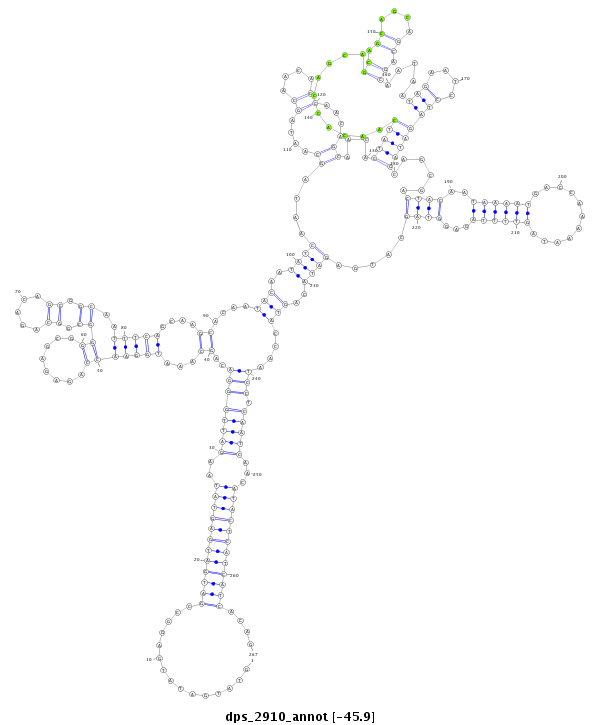 |
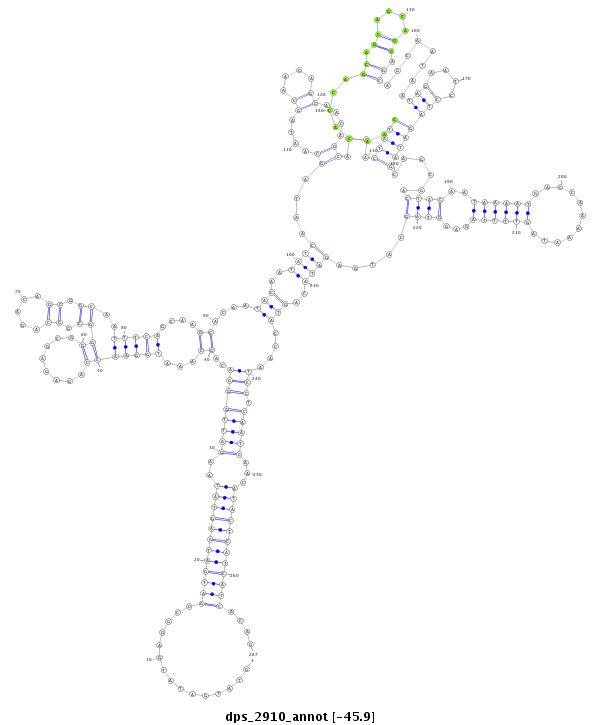 |
CDS [Dpse\GA12452-cds]; exon [dpse_GLEANR_1453:4]; CDS [Dpse\GA12452-cds]; exon [dpse_GLEANR_1453:3]; intron [Dpse\GA12452-in]
| Name | Class | Family | Strand |
| (CTG)n | Simple_repeat | Simple_repeat | + |
| TART_DV | LINE | Jockey | - |
| ##################################################---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTCCAGAGCGCAAGCAGCAACAGCAGCAGCAGCAGCAGCAACAGAACGATGTATGATATGAGGCCGATGATGAGTATAAGATTGGGACAGCAAATGGAACCAGAGAGCGGGGCGGCAGACAGCGGCAATTTCAGCAAGCACAATACAATATCAATAGCAATAGCAACAGCAACAGCAACACTATCAACACCAGCAGCAGCAGCAGCAGCAATAATAGAATCCTAGATAGCAGCGACTACAATAAAATGACCAAAAATAGTTTTAGAGGTAGCATGAGATACAGTACCAATCCTCAATCAACATACTCATCATCACAGCAGCAGAAGCAGCAGCAGCAGCAGAAAGAGAAGCACCATCAGCAACATAA **************************************************...............((((((((((((..((((((((..((...(((((((........))((.((.....)).))..)))))....))....(((..((((....((....))....((....))....(((((.......((.((....)).))........(((....))))))))......((((..((((((...........))))))...)))).....))))..)))....))).)))))...))))))))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
GSM444067 head |
V112 male body |
M059 embryo |
SRR902010 ovaries |
SRR902011 testis |
M062 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................AACAACTACACTATCATCACC................................................................................................................................................................................ | 21 | 3 | 1 | 23.00 | 23 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................ACAACTACACTATCATCACC................................................................................................................................................................................ | 20 | 3 | 10 | 1.20 | 12 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AGCGGGGCGACAGACAGC.................................................................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TATCGACACCAGCAGCAGCAGCA................................................................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................AACAACTACACTATCATCAC................................................................................................................................................................................. | 20 | 3 | 16 | 0.81 | 13 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................ACAACTACACTATCATCACCA............................................................................................................................................................................... | 21 | 3 | 7 | 0.71 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................GAGCCAGAGAGCGGGGCGGCT.......................................................................................................................................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TGGGGCGGGAGACAGCGGCGAT.............................................................................................................................................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................ATGATGAGTATCAGGTTG........................................................................................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................CAGCGGCAAATTCAGTAAG..................................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................GCAGCAGCAGCAAGAGAAGCACC............. | 23 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CGCTAGCAGCAACAGCAGCAGC................................................................................................................................................................................................................................................................................................................................................. | 22 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TCAACACTAGCAGCAGCAGCA................................................................................................................................................................... | 21 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........GCAAGCAGCGACAGCAGCA................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................CAACACCAGCAGAAGCAGCAGCAGC.............................................................................................................................................................. | 25 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................TAGAAGTAGCAGCAGCAGGAGAAA....................... | 24 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ...................................................................................................................................................................................................................................................................................................................ATCATCGAAGCAGCAGAAGC........................................ | 20 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................GCAGCGGCAATTTCGGCGAG..................................................................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............GCAGCAACAGCAGCAGCAGCAGCAGCAAC..................................................................................................................................................................................................................................................................................................................................... | 29 | 0 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................CAACACCAGCAGCAGCAGC.................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................TGGTGAGTGTAAGACTGGG......................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................CACCAGCATCAGCAGCAGCAGCAA............................................................................................................................................................ | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AAGGTCTCGCGTTCGTCGTTGTCGTCGTCGTCGTCGTCGTTGTCTTGCTACATACTATACTCCGGCTACTACTCATATTCTAACCCTGTCGTTTACCTTGGTCTCTCGCCCCGCCGTCTGTCGCCGTTAAAGTCGTTCGTGTTATGTTATAGTTATCGTTATCGTTGTCGTTGTCGTTGTGATAGTTGTGGTCGTCGTCGTCGTCGTCGTTATTATCTTAGGATCTATCGTCGCTGATGTTATTTTACTGGTTTTTATCAAAATCTCCATCGTACTCTATGTCATGGTTAGGAGTTAGTTGTATGAGTAGTAGTGTCGTCGTCTTCGTCGTCGTCGTCGTCTTTCTCTTCGTGGTAGTCGTTGTATT
**************************************************...............((((((((((((..((((((((..((...(((((((........))((.((.....)).))..)))))....))....(((..((((....((....))....((....))....(((((.......((.((....)).))........(((....))))))))......((((..((((((...........))))))...)))).....))))..)))....))).)))))...))))))))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M062 head |
SRR902010 ovaries |
GSM343916 embryo |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................TTGTTGATGTGATAGTAGTGG................................................................................................................................................................................ | 21 | 3 | 1 | 107.00 | 107 | 73 | 34 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................CTTCGTGGTAGTCATTGTTT. | 20 | 2 | 3 | 9.00 | 27 | 0 | 0 | 27 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TGTTGATGTGATAGTAGTGGT............................................................................................................................................................................... | 21 | 3 | 7 | 4.14 | 29 | 7 | 22 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TTGTTGATGTGATAGTAGTG................................................................................................................................................................................. | 20 | 3 | 16 | 3.00 | 48 | 38 | 10 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TTGTTGATGTGATAGTAGTGGT............................................................................................................................................................................... | 22 | 3 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TGTTGATGTGATAGTAGTGG................................................................................................................................................................................ | 20 | 3 | 10 | 2.70 | 27 | 17 | 10 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................CTTCGTGGTAGTCATTGTGT. | 20 | 2 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................TCTCTTCGTGGTAGTCGTTGTAT. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................TCGTGGTAGTCATTGTAT. | 18 | 1 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................TTCGTGGTAGTCGTTGTGT. | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................ATGTCATGGTTAGGAGTTAGTTGT................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................TTTCGTGGTAGTCATTGTAT. | 20 | 2 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................TCTTTATCAGAATCTCAATCGT.............................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................CGTCGTGGTAGTCATTGTAT. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..GGTCTCGCGTGCGATGTTGT......................................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................ACTTGGTTCGGAGTTAGTTG.................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................GTGAAAGTTGTGGCCGTCGA......................................................................................................................................................................... | 20 | 3 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................AGCCTGTCGTTTACCTCGGTG........................................................................................................................................................................................................................................................................ | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................GTAGTTGTCGCTGTGATAG...................................................................................................................................................................................... | 19 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................TTCGTGGTAGTCATTGTCT. | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................GTTGTTGATGTGATAGTAGTG................................................................................................................................................................................. | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................CGGTTGTCGTTGTGATAGTTG................................................................................................................................................................................... | 21 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................GTGAAAGTTGTGGCCGTC........................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........GTTCGTCGTTGTCGTCGTC.................................................................................................................................................................................................................................................................................................................................................. | 19 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TAGTTGTGGTCGTCGTAATCGT................................................................................................................................................................... | 22 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................GACGTCTTAGTCGTTGTCTTGC............................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................CTTCGTGGTAGTCATTGCTT. | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................TCTTTGTGATAGTTGGGGA............................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................CTTCGTGGTAGTCATTTTTT. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................GTTGTCGTTGTCGTTGTG.......................................................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GTTGTGGTCGTCTTCGTCGTCG................................................................................................................................................................. | 22 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............GTCGTTGTCGTCGTCGTCG.............................................................................................................................................................................................................................................................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .GGAACTCGCGTTCGTCGTT........................................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................CTTCTTGGTAGTCATTGTTT. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group3a:1194520-1194886 - | dps_2910 | TTCCAGAGCG-CAAGCAGCAACAGCAGCAGCAGCAGCAGCAACAGAACGATGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ATGGAACCA---------------------------------------------------------------------GAGAGCGGGGCGGCAGACAGCGGCAATTTCAGCAAGCACA---ATACAATATCAATAGCAA---TAGC---AACAGC---------------------------AACAGCA---------------ACAC---------------TAT--------CAACACCA---------GCAGCAGCAGCAGCAGCAATA---ATA------------------GAATCCTAGATAGCAG---------CGACTACAATAAAATGACCAAAAATAGTTTTAGAGGTAGCATGAGATAC---------AGTACCAATC---CTCAATCAACATACTCATCATC------------ACAGCAGCAGAAGCAGCAGCAGCAG------CAGAAA---------GAGAAGCACCA---------TCAGCAACATAA |
| droPer2 | scaffold_23:1215162-1215539 - | dpe_1209 | TTCCAGAGCG-CAAGCAGCAACAGCAGCAGCAGCAGCAGCAACAGAACGATGTATGATATGAGGCCCGATGATGAGTATAAGATTTGGGACAGCAA------------------ATGGAACCA---------------------------------------------------------------------GAGAGCGGGGCGGCAGACAGCGGCAATTTCAGCAAGCACA---ATACAATATCAATAGCAA---CAGC---AACAGCA------A---------CAG---CAACAGCAACA------------------C---------------TAT--------CAACACCAGC------AGCAGCAGCAGCAGCAGCAGCAATAACA------------------GAATCCTAGATAGCAG---------CGACTACAATAAAATGACCAAAAATAGTTTTAGAGGTAGCATGAGATAC---------AGTACCAATC---CTCAATCAACATACTCATCATC------------ACAGCAGCAGAAGCAGCAGC---AG------CAGAAA---------GAGAAGCACCA---------TCAGCAACATAA |
| droWil2 | scf2_1100000004540:132937-133339 - | dwi_1393 | TTCCCGAGCG----------------------TAAACAGCAGCAGAATGATGTATGATATAAGGC-TGATGATGAGTATAAAAT-TGGGATAACAAATGCAACAGCAGCAAC------AAGCA---------------------------------------------------------------------GACAACGAGGCAGCAGACAGCGACAATTTC-----------------T------AAGACAA---CAAC---AACAGCA------GCAGCAA------CAGCAAT----------CCAGCAACAGCAACAGCAATAACAACAACAACAATAATCA--CAACAGCAACACA------------------AGCAAT---CACA---------------ATCAAATCGTAGATAGCACTGCAACAACAAACTACAATAAAATGACCAAAAATAGTTTTAGAGGACAGATGAGATTTACCACACCAAACTCCA------CTCAATTGTCATACTCATCATCATTATCATCATCACAACAACAGCAACAG---C---AA------AAGCCGAAGCAGAAAC---------AGAGACAGCAACAGCAATATAG |
| droVir3 | scaffold_13049:7802144-7802528 - | dvi_18894 | TACCAGAGCG-CAGCCAGCAG------------CTGCAGCAGCAGGATGAGGTATGATATAAGGC-TGATGATGAGTATAAGAT-TGGGACAGCAC------------------ACACAACGG---------------------------------------------CGCCGGCAGCGGAGGCAGTGAATCGGCAACGAGGCGGCAGACAGCGACAGTTTCTG---GCACA---AAACA------AAAGCAA---CATC---AACAGCA------G---------CAGCAGCAGCAGCAGCA---------------ACATAGATA---------AAAGCAAGCAC--------------------------ATCAACAACAACA---ACA------------------GAATCGTAGATAGCACCGCACCAACCAATTACAATAAAATGACCAAAAATAGTTTTAGAGGCAGCATGAGAT-----ACACAAACAATCAAT----CTCAATCGTCATACTCATCATC------------ACAGCAGCAACAGCAACAGC---AA------CAGCAG---------------CAGCAAC------AGCAACAATATAA |
| droMoj3 | scaffold_6680:11451832-11452198 - | TACCGGAGCG-CAGCCAACAGCAGCAGCAGCAGCAGCAGGAGCAGGCTGAGGTATGATATAAGGC-TGATGATGAGTATAAGAT-TGGGAAAACAC------------------ACACAACAA---------------------------------------------CG------GCGGAGGCAGCGGATCGGCAGCGAGGCAGCAGACAGCGACAGTTTCTG---GCACA---AAACA------AAAGCAG---CAGC---------A------G------------CAGCAGCAACAGCA---------------ACATAGATA---------AAAGCAAA--CACAACAGCAAT------------------AGCAACAACA---ACA------------------GAATCGTAGATAGCACCTCACCAACCAACTACAGTAAAATGACCAAAAATAGTTTTAGAGGCAGCATGAGATAC--AAA-------ACCAATCAATCTCAATCGTCATACTCATCATC------------ACAGCAACAGCAACAG---------------------------------------------------CAACAATTTAA | |
| droGri2 | scaffold_15110:6855400-6855763 - | TACCGGAGCG-TAATCAACAG------------CAGCAGCAGCAGGATGAGGTATGATATAAGGC-TGATGATGAGTATAAGAT-TGGGACAGCGA----------------------AAC-----------------------------------------------------------------------GACAACGAGGTGGCAGACAGCGTCAGTTTGTG---GCACAGCAAAACA------CAAGGAACATCAGC---AACATCA------GCAACAACAGCAGCAGGAGCAGCAACA---------------ATATAGATAGCAA---------CCAGC-----ACAACAAT------------------AAGAACAGCA---ACA------------------GGATCGTAGATAGCACCGCAAAAATTAACTACAATAAAATGACCAAAAATAGTTTTAGAGGCAGCATGAGATACAC------A---AGCAATCCATCTCAGTCGTCATACTCATCATC------------ACAGCAGCAACAGCAACAACAGCAA---------------------C------------------AGCAACAATATAA | |
| droAna3 | scaffold_13337:13457239-13457590 + | TCCCGGAGCG----------------------CCAACAGGCGGCTAATGATGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGCAGACAGCGGCAATTTC-----------------A------ACAGCAA---CAAC---AGC------------------------------AGCAGCAACATCAGCAACAGCAAC------------AGCAACAGCAATCACTCAACAACAATAT------------------AAGCAGCA---ACA------------------GAATCGTAGATAGCAG---------CAACTACAATAAAATGACCAAAAATAGTTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAGCAACAGCAACAGC---AA------CAGCAA---------CAGCAGCA---TCAGCAGTATCAGCAACATAA | |
| droBip1 | scf7180000396589:558308-558659 - | TCCCCGAGCG----------------------CCAACAGGCGGCTAATGATGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGCAGACAGCGGCAATTTC-----------------A------AAAGCAA---CAAC---AGCAGCA------A---------------------CAGCAACATCAGCAACAGCAACATCAGCA---------ACAGCAATCACTCAACAACAATAT------------------AAGCAGCA---ACA------------------GAATCGTAGATAGCAG---------CAACTACAATAAAATGACCAAAAATAGTTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAGCAACAGCAACAGC---AG------CAGCAA---------C---------ATCAGCAGTATCAGCAACATAA | |
| droKik1 | scf7180000302274:515144-515471 - | TCCCCGAGAA----------------------GCAGCAGTCGTCGAGTGATGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCA------------GCAGCAAACGGAACCACAGC---------------------------------------------------------------ACAGCAGCGCGGAGGCAGACAGCGGCAATTTC-----------------A------AAAGGAGAAACAGC---AACAGC---------------------------AGCAGCA---------------------------------ACATCA---ACTCAAC---------------------------AGCAACA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCGTACTCATCATC------------ACAGCAGCAGCAGCAACAGG---AG------CAGCCA---------CAG------------CAGAAACAGCAACATAA | |
| droFic1 | scf7180000454065:475116-475464 - | TCCCGGAGCG----------------------CCAGCAGCAGCTGAATGAAGTATGATATGAGGC-CGGTGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAAACAGCGGCAATTTC-----------------A------AAAAGAA---CAAC---AACAGCA------GCAACAG------CAGCAACAACAGCA---------------ACAACAACAGCAACAGCAACAACAACCACTAAACAGCAAC------ATCAGCGGAATAAATAGCAGCA---ATA------------------GAATTGTAGATAGCAG---------CAGCTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCTTCATACTCATCATC------------ACAACAACAGCAGA---------------------------------------------------AGCAGCAACATAA | |
| droEle1 | scf7180000491249:6079420-6079786 + | TCCCAGAGCG----------------------CCAGCAGCAGAAGAATGAAGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAGACATCGGCAATTTC-----------------A------AAAGCAA---CAGC---AACAGCA------G---------CAGCAG---CAGCTGCA---------------ATATCAACAGCAGCAACATCAGCTACAACTCAACAGCAAC------ATCAGCGGAATTAATAGCAACA---ATA------------------GAATCGTAGATAGCAG---------CGACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAGCAGCAGCAACAGC---AA------CAGCAA---------C---------ATCAGCATCAAAAGCAACATAA | |
| droRho1 | scf7180000779841:94293-94642 + | TTTCAGATCTGGGAGCAACAGCAACATCAACAGCAACAGC------------------------------------------------------------------------------AACCACAGCGGAGCCTGATATTGATGGCATGTCGACAACAACACATGCAGCA----------------------GCCATGGCA-------ACAGCAACAACTAC-----------------A------TAAAAAA---CAGC---AGCAGCAACTGTAT---------CAGGAGCAACAGCAGCA---------------------------------GCATCAA-----CTGCAGCAACAGTACCATCAGCAACAGCATCAGCAGTA---CCAAAAACAACAGCAGCA---GCAAC---ACTAACAG---------CAACACCAACAGCAACACCAAAAGCAGCAGCAGCAACAGCAGCAGCAAT---ATACAGATTGCCAA-------------------------CA------------ACAGCAGCAACATCATCGGC---AG------CAGCAG---------C---AACAA------CATGAGTGACAACAACA | |
| droBia1 | scf7180000302428:1095420-1095774 + | TCCCAGAGCG----------------------CCAGCAGCAGCTGAATGAAGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAGACAGCGGCAATTTC-----------------A------AAAGCAA---CAGC---AACAGCA------G---------C---------AGCAGCA---------------------------------ACATCAGGCACTCAACAGCAACTCGGCAATCGTCGGAATCAACAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAATAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAGCAACAGCAGCAACAGCAG---------------------CAGCAGCAACATCGGAATCAGAAGCAACATAA | |
| droTak1 | scf7180000415862:231863-232217 - | TCCCGGAGCG----------------------CCAGCAGCAGCAGAATGAAGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------CACAGCGCGGCGGAAGACAGCGGCAATTTC-----------------A------AAACCAA---CAGC---CACAGC------------------------AACAGCAGCA---------------------------------ACATCAAGCAATCAACAGCAACTCGGCAATCAGCGGAATCAACAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAATAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAGCAGCAGCAGCAACAGCAA---------------------CAGCAGCAACATCAGAACCAGAAGCAACATAA | |
| droEug1 | scf7180000409524:487186-487507 + | TCCCAGAGCG----------------------CCAGCAGCAGCTGAATGAAGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAGACAGCGGCAATTTC-----------------A------AAAACAA---CAGC---A---GC---------------------------AACAGCA---------------------------------ACATCAAGCACTCAACAGCAAC------ATCAGCGGAATTAATAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAACAGCAACAGCAGC---AG------C---------------------------------AGCAGCAACATAA | |
| dm3 | chr3L:8153940-8154300 - | TCCCAGAGCG----------------------CCAGCAGCAGCTGAATGATGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGGGGCGGAAGACAGCGGCAATTTC-----------------A------AAAGCAA---CAGC---AACAGAA------G---------CAG---CAACAGCAGCA---------------------------------ACATCAAGCACTCAACAGCAACACGGCAATCGGCGGAATCAACAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CCCAATCGTCATACTCATCATC------------ACAGCAGCAACAGCAGCAGC---AACAACAACAA------------CAGAAGCAACATC------AGAAGCAACATAA | |
| droSim2 | 3l:7963658-7964015 - | dsi_20976 | TCCCAGAGCG----------------------CCAGCAGCAGCTGAATGATGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAGACAGCGGCAATTTC-----------------A------AAAGCAA---CAGC---AACAGCA------G---------CAGCAGCAACAGCAGCA---------------------------------ACATCAAGCACTCAACAGCAACACGGCAATCGGCGGAATAAACAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CCCAATCGTCATACTCATCATC------------ACAGCAGCAGCAGCAACAAC---AA------CAA------------CAGAAGCAACATC------AGAAGCAACATAA |
| droSec2 | scaffold_0:440983-441349 - | TCCCAGAGCG----------------------CCAGCAGCAGCTGAATGATGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAGACAGCGGCAATTTC-----------------A------AAAGCAA---CAGC---AACAGCA------G---------CAGCAGAAACAGCAGCA---------------------------------ACATCAAGCACTCAACAGCAACACGGCAATCGGCGGGATAAACAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CCCAATCGTCATACTCATCATC------------ACAGCAGCAGCAGCAACAACAGCAACAACAACAA------------CAGAAGCAACATC------AGAAGCAACATAA | |
| droYak3 | 3L:8742441-8742816 - | TCCCAGAGCG----------------------CCAGCAGCAGCAGAATGAAGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAGACAGCGGCAATTTC-----------------A------AAAGCAA---CAGCACCAACAGCA------G---------CAGCAGCAACTG------------------CAACAGCAGCAAC---AGCAACATCAAGCAATCAACAGCAACACGGCAATCGGCGGAATCAACAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAGCAGCAGCAACAGC---AA------CAGCAA---------CACAAGCAACATC------AGAAGCAACACAA | |
| droEre2 | scaffold_4784:3208791-3209127 + | TCCCAGAGCG----------------------CCAGCAGCAGCCGAATGAAGTATGATATGAGGC-CGATGATGAGTATAAGAT-TGGGACAGCAA------------------ACGGAACCA---------------------------------------------------------------------GACAGCGCGGCGGAAGACAGCGGCAATTTC-----------------A------AAAGCAA---CAGC------------------------------------AGCAGCA---------------------------------ACATCAAGCACTCAACAGCAACACGGCAATCGGCGGAATCAACAGCAGCA---ATA------------------GAATCGTAGATAGCAG---------CAACTACAGTAAAATGACCAAAAATAGCTTTAGAGGTAGCATGAGATTTACCACACCAAGTACCAATC---CTCAATCGTCATACTCATCATC------------ACAGCAGCAGCAGCAACAAC---AA------C---------------ACAAGCAACAGC------AGAAGCAACACAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 05:47 AM