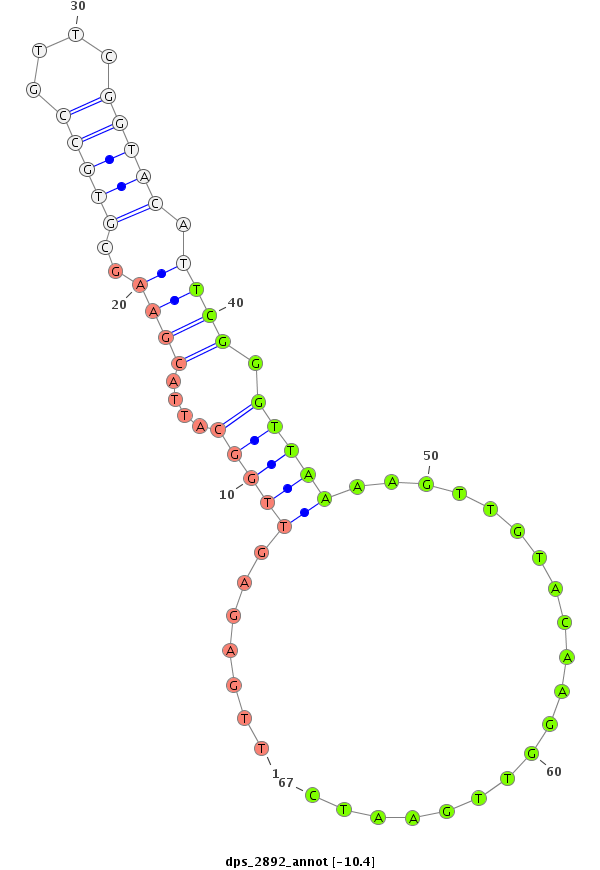
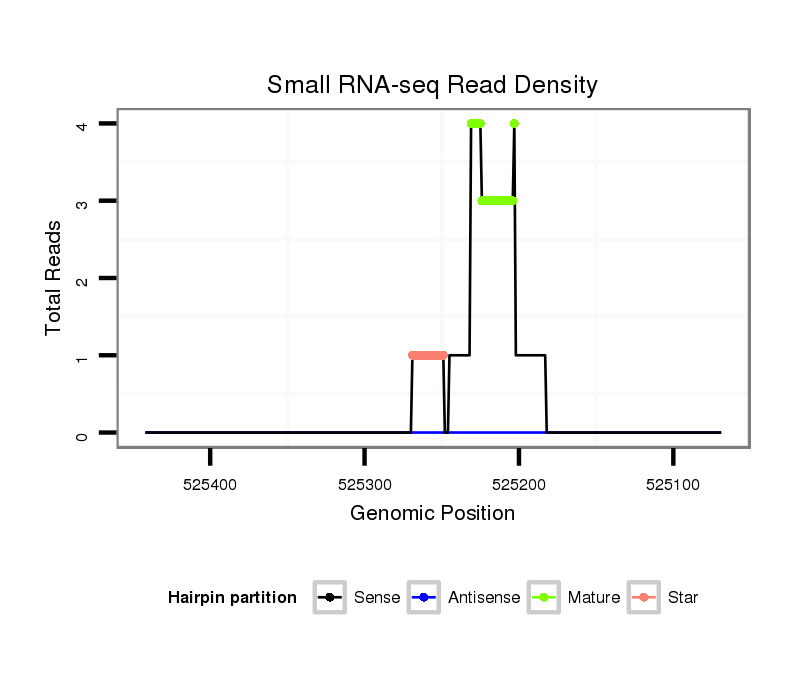
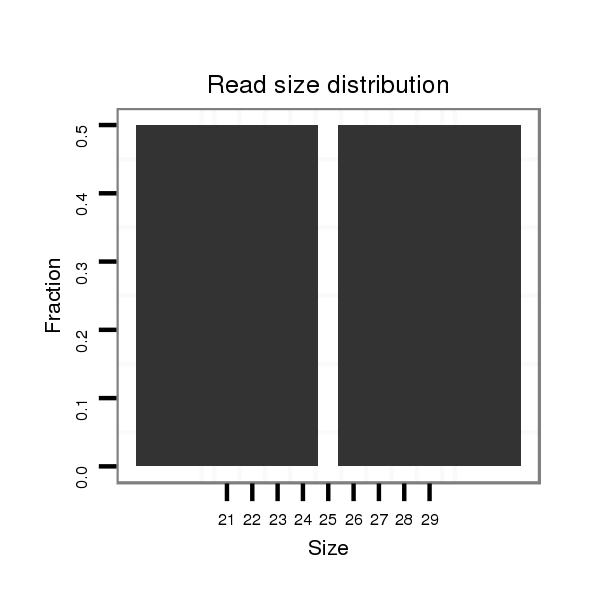
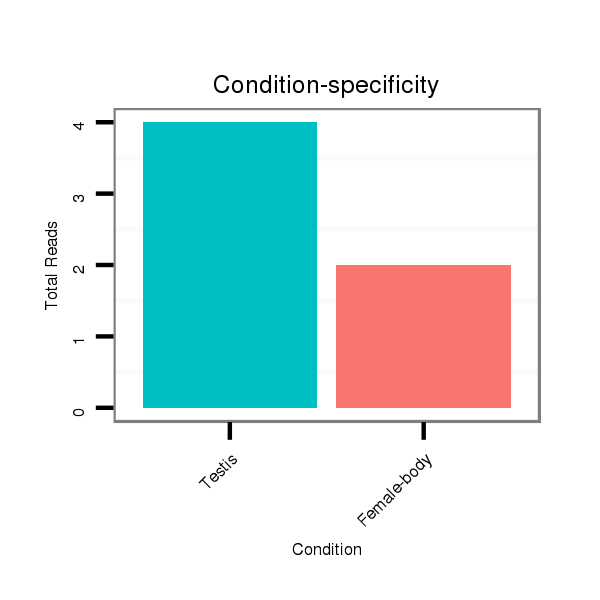
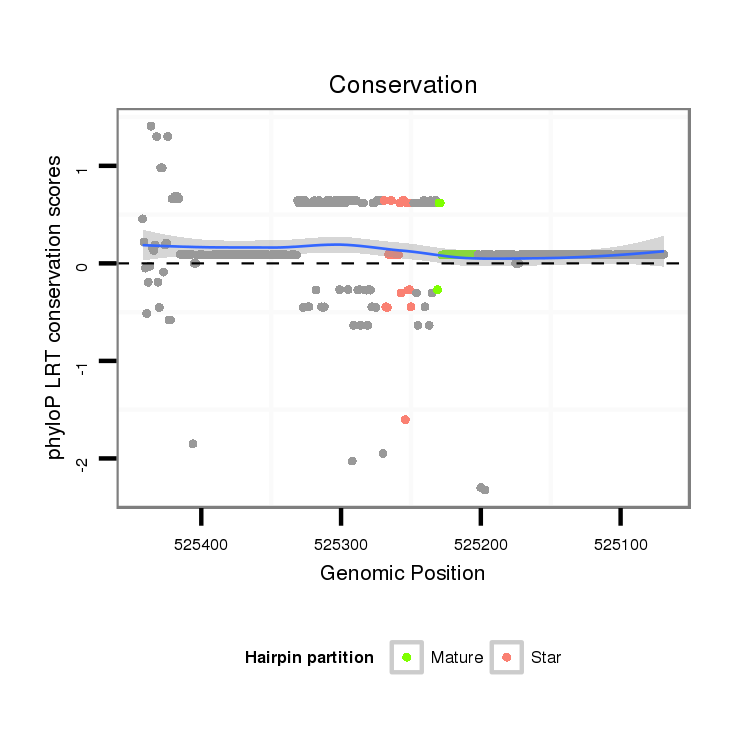
ID:dps_2892 |
Coordinate:XR_group3a:525119-525392 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -10.6 | -10.3 | -10.1 |
 |
 |
 |
exon [dpse_GLEANR_1486:2]; CDS [Dpse\GA24527-cds]; CDS [Dpse\GA24527-cds]; exon [dpse_GLEANR_1486:1]; intron [Dpse\GA24527-in]
No Repeatable elements found
| mature | star |
| ##################################################----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------########################-------------------------- CGCTCCGTTTTACGGACAACCCGAATGAGCTAGTCAACACCATGCGCGCGGTGTTTCTAATAAACCAGATAAGTATTTGGAACAACTGCAGCCATTCAGAAATCCGCTTGGAGGTCTCATGCGACCAGACTCACTGGGATCATGTGAATATGATCAGCGCCGTCTGCCATATTTTGAGAGTTGGCATTACGAAGCGTGCCGTTCGGTACATTCGGGTTAAAAGTTGTACAAGGTTGAATCTCGGTCTGATGGCCAGGTTGATCAGAGCCCCTGAATGCAGTTCACTTCTGACAACTCTTCAAATATAGTTTCCAATGTTTAAAGAACAACAATTTGGGCACCAGCTAATAGGTGTATATGCTGATACGAATAAA *****************************************************************************************************************************************************************************.......(((((....((((..(((((....))))).)))).)))))....................************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
GSM444067 head |
M059 embryo |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................TCGGGTTAAAAGTTGTACAAGGTTGAATC...................................................................................................................................... | 29 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................CTCGGTCTGATGGCCAGGTTG.................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................ATTTGGCACACCTGTAGCCATT...................................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................TTGAGAGTTGGCATTACGAAG.................................................................................................................................................................................... | 21 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................GCCGTTCGGTACATTCGGGTT............................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................ATGCGCGGGGTGTTTCGA........................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................TTAAAGAACAACAATTTGGG.................................... | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TGCGCGCGGGGTTTCAAAAAAA...................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........TACGGACAACCCGAATGAGCT....................................................................................................................................................................................................................................................................................................................................................... | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................TCAACGTCATGCGCGCGGCG................................................................................................................................................................................................................................................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ATGTGAATATGCTCAGCGC...................................................................................................................................................................................................................... | 19 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AAAAATGATCAGCACCGTCT................................................................................................................................................................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................GGACAACAATTTGGGGAC................................. | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................GGGGGGCTCATGCGAACAG...................................................................................................................................................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TGCGCGCGGGGTTTCAAAAA........................................................................................................................................................................................................................................................................................................................ | 20 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CGTACGGTTTAAAAGTTGT................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................AAAATGATCAGCACCGTTT................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................CACCAGAATTAGCTAGTC................................................................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GCGAGGCAAAATGCCTGTTGGGCTTACTCGATCAGTTGTGGTACGCGCGCCACAAAGATTATTTGGTCTATTCATAAACCTTGTTGACGTCGGTAAGTCTTTAGGCGAACCTCCAGAGTACGCTGGTCTGAGTGACCCTAGTACACTTATACTAGTCGCGGCAGACGGTATAAAACTCTCAACCGTAATGCTTCGCACGGCAAGCCATGTAAGCCCAATTTTCAACATGTTCCAACTTAGAGCCAGACTACCGGTCCAACTAGTCTCGGGGACTTACGTCAAGTGAAGACTGTTGAGAAGTTTATATCAAAGGTTACAAATTTCTTGTTGTTAAACCCGTGGTCGATTATCCACATATACGACTATGCTTATTT
**************************************************************************************************************************************.......(((((....((((..(((((....))))).)))).)))))....................***************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
M022 male body |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|
| .............................................CGCGTCCCAAAGATTATCTGG.................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 2 | 3.50 | 7 | 7 | 0 | 0 | 0 |
| .............................................CGCGTCCCAAAGATTATCTG..................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 4 | 3.00 | 12 | 10 | 2 | 0 | 0 |
| .............................................CGCGTCCCAAAGATTATCTGGTC.................................................................................................................................................................................................................................................................................................................. | 23 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| .............................................CGCGTCCCAAAGATTATCTGGT................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ...............................................CGTCCCAAAGATTATCTGGTC.................................................................................................................................................................................................................................................................................................................. | 21 | 3 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................ACCGAAAAGCGTCGCACGG.............................................................................................................................................................................. | 19 | 3 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 |
| ...............................................CGTCCCAAAGATTATCTGG.................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.30 | 3 | 3 | 0 | 0 | 0 |
| .............................................................................................TAAGTGTTTAGTCGAACCG...................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.25 | 5 | 4 | 0 | 0 | 1 |
| .............................................................................................TAAGTGTTTAGTCGAACC....................................................................................................................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ........................AACTCGGTCAGTTGTGGT............................................................................................................................................................................................................................................................................................................................................ | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................TAAAGTGGAGACTGTTGA.............................................................................. | 18 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................CGGTTATACTAGTCCCGGC.................................................................................................................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................CTTAAAGTGGAGACTGTTGA.............................................................................. | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................CTTAAAGTGGAGACTGTTG............................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................CTGGAGGACTTACGTTAAG........................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................CCCAAAGATTATCTGGTCA................................................................................................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ....................................................CAAAGATTATCTGGTCACT............................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XR_group3a:525069-525442 - | dps_2892 | CGCTCCG---TTTTACGGACAACCCGAATGAGCTAGTCAACACCA--TGCGCGCGGTGTTTCTAATAAACCAGATAAGTATTTGGAA---CAACTGCAGCCATTCAGAAATCCGCTTGGAGGTCTCATGCGACCAGACTCACTGGGATCATGTG-------------AATATGATCAGCGCCGTCTGCCATATTTTGAGAGTTGGCATTACGAAGCGTGCCGTTCGGTACATTCGGGTTAAAAGTTGTACAAGGTTGAATCTCGGTCTGATGGCCAGGTTGATCAGAGCCCCTGAATGCAGTTCACTTCTGACAACTCTTCAAATATAGTTTCCAATGTTTAAAGAACAACAATTTGGGCACCAGCTAATAGGTGTATATGCTGATACGAATAAA |
| droPer2 | scaffold_23:542851-543224 - | CGCTCCG---TTTTACGGACAACCCGAATGAGCTAGTCAT--CCAGCTGCGCGCGGTGTTTCTAATAAACCAGATAAGTATTTGGAACTACAACTGCAGCCATTCAGAAATCCGCTTGGAGGTCTCATGCGACCAGACTCACTGGGATCATGTG-------------AATAAGATCAGCGCCGTCTGCCATATGTTGAGAGTTGGCATTGCGAAGCGTGCCGTTCGGTACATTCGGGTTAAAAGTTGTACAAGGTTGAATCTCCGTGTGATGGCCAGGTTGATCAGAG---CTGAATGCAGTTCACTTCTGACAACTCTTCAAATATAGTTTCCAATGTTTAAAGAACAACAATTTGGGCACCAGCTAATAGGTGTATATGCTGATACGAATAAA | |
| droWil2 | scf2_1100000004921:3087019-3087037 + | CACAATG---CACTAAGGGTAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15081:2616477-2616501 + | CATCGCGTTGCATT-----CAAGGCGAATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302517:142690-142801 + | TGG--------------------------------------------------------------------------------------------------------------------AGGTATCCGGCGATCAGCAGCACTGGGATCGTGTGGTGGACTACAGTGAGTACCATTGCCGTTCCTGGCAATATTTGT--A-----CTTTGCAGCGCGACCCGTGCGCTTCATCCG---------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000302126:3594754-3594772 + | CGCGCTG---CATTCAGGGCGA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSim2 | x:10121949-10121955 + | CGCGCTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_8:1140439-1140445 + | CGCGCTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4690:4467192-4467198 - | CGCGCTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 05:43 AM