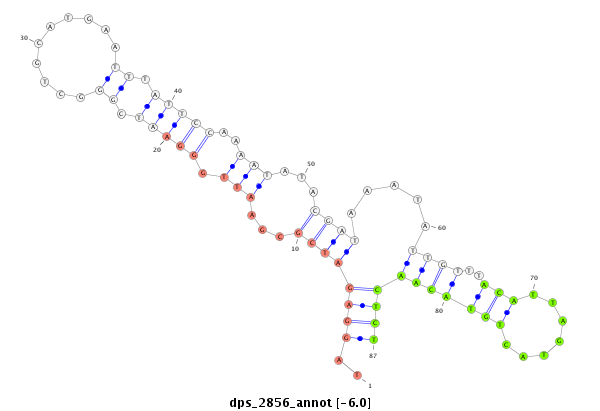
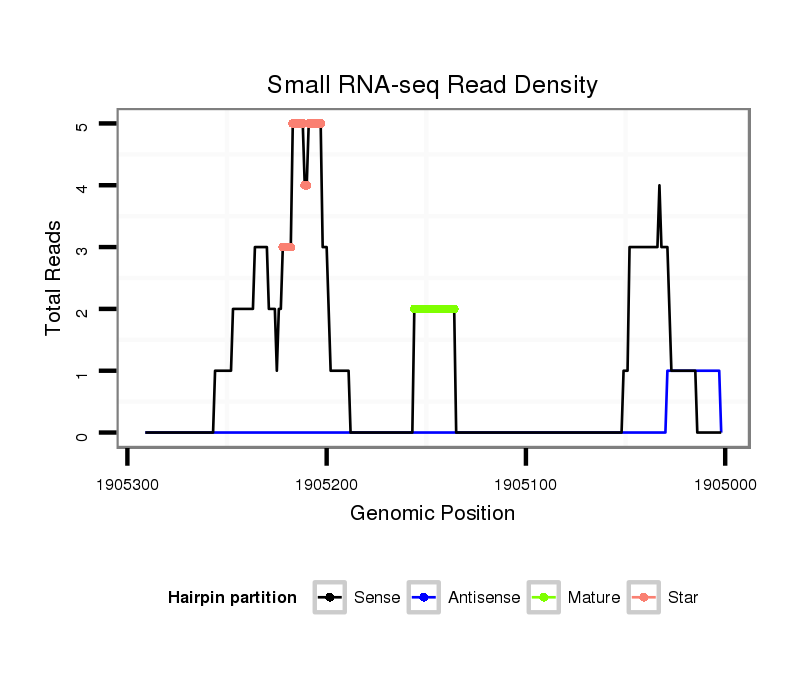
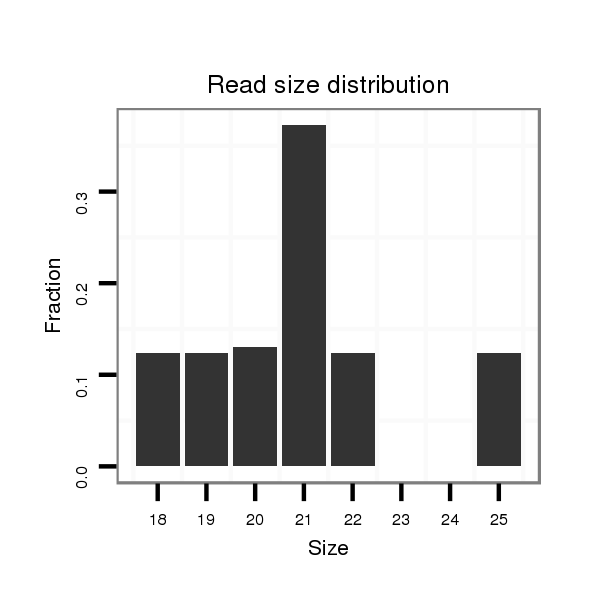
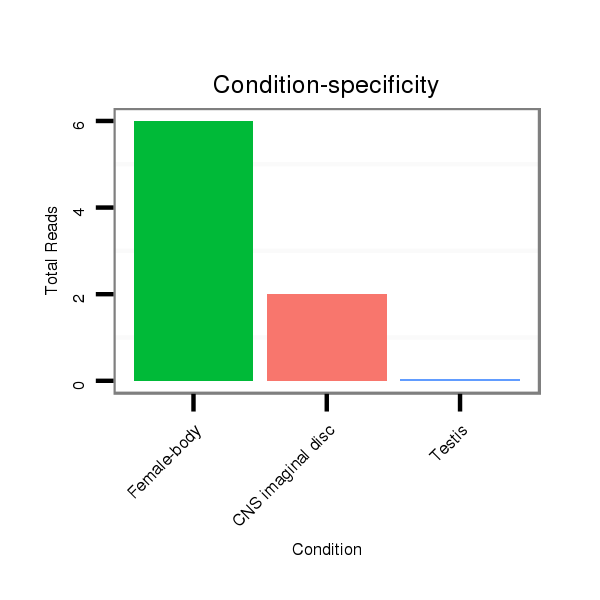
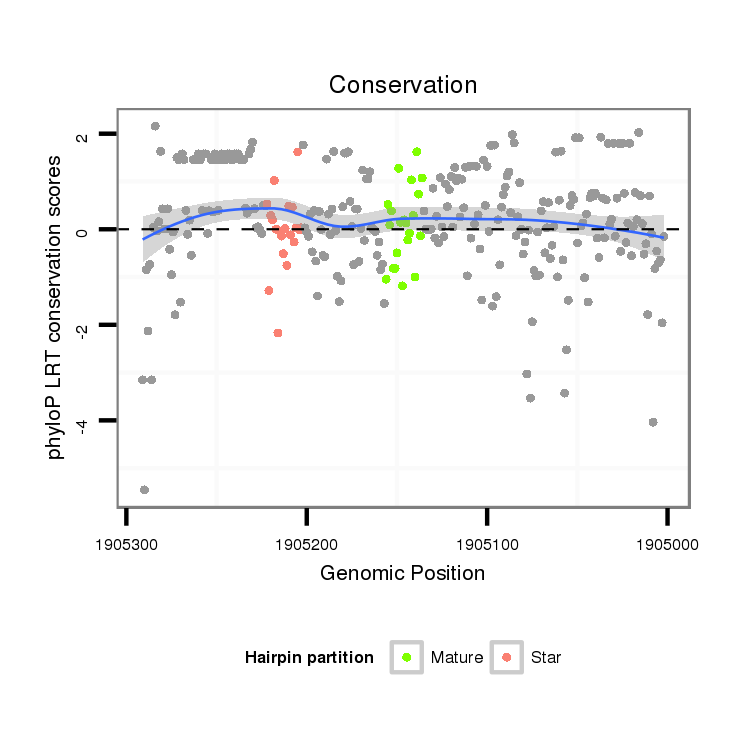
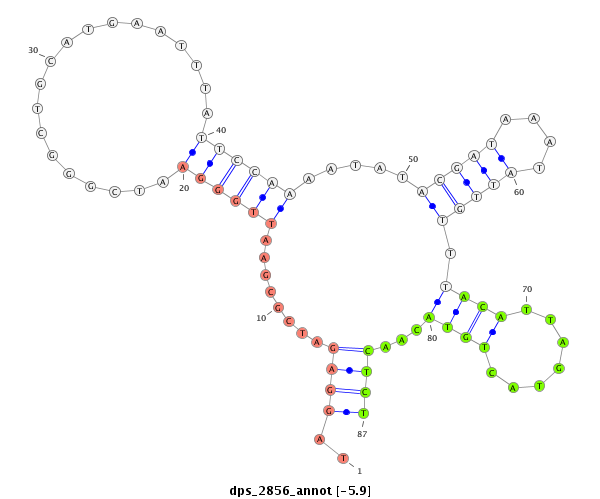
| dp5 |
XL_group3a:1905002-1905291 - |
dps_2856 |
CACGGAGAAGTGCG-CCAATGG-TACCGTGGCCTGCTATTCTTGCGAGCA-AGGTTAG-G------GTGAGTGGGGAAA----TAGGAGATCGCGAATTGGGAATC-G-GGCTGCAT----------------------------GAATTTATT------------------------------------------------------------------------------------CCAA----------------------------------AA--------TATACGATAAATATT--------GT--------------------------------------------------TTACA---------TTAGTACTGTACA----A--CTCT----------CTCTCTCTG--TCTCT--CTC--TCTCT--GT------GTCTCTCTTT----CTCTCTCTG----TC--TCTCTCA----CTCTGATG--------------------------------------TCCTTT------C-----------------------------CTTCTTT-----T-------------TG--------------------AGGGCAGATACTGTCCTGCGACGTGGATGCGCCGATG--CCGACTT--CTCACCCTCCGAC------------------------- |
| droPer2 |
scaffold_11:2774738-2775023 + |
|
CACGGAGAAGTGCGTCCAATGGTAACCGTGGCCTGCTATTCTTGCGAGCAAAGGTTAGAG------GTGAGTGGGGAAA----TAGGAGCTCGCGAATTGGGAATC-G-GGCTGCAT----------------------------GAATTTATT------------------------------------------------------------------------------------CCAA----------------------------------AA--------TATACGATAAATATT--------GT--------------------------------------------------TTACA---------TTAGTACTGTACA----A--CTCT----------C----------TCTCT--CTC--TCTCT--GT------GTCTCTCTTT----CTCTCTCTG----TC--TCTCTCA----CTCTGATG--------------------------------------TCCTTT------C-----------------------------CTTCTTT-----T-------------TG--------------------ACGGCAGATACTGTCCTGCGACGTGGATGCGCCGATG--CCGACTT--CTCACCCTCCGAC------------------------- |
| droWil2 |
scf2_1100000004909:11942267-11942508 - |
|
CACGGAGAAGTGTG-CTGCCGG-CACCGTTGCCTGTTATGCCTGTGAGCA-AGGTTAG-G------GTGAGTCATCCAA----A-------------------------------------------ATAAA-G---------TG---------TATGTG---------TGTGTT------AGTCAAAATCAATAAA---------------------------------------------------------------------------------------------------------AAAAAAAAAAAACTCTAATCAAACTCGAGTGGTTTTTCTGTTTCA--TTC--AATCTCTAATCTC-----------------------------------------------------------------T--GC--CT--AAATATGTAT--------ATT-----------------------------------------------------------------------------------------------------------------------------------------------------------------CCTCCAGATACCTTTCTTCGTCGCGGCTGTGCCGATG--CTGATTT--CATCCCAGCCACT------------------------- |
| droVir3 |
scaffold_12928:5004347-5004471 + |
|
TGA-------------------------------------------------------------------GTGCGGAAA----TCA--------AAATAGGAAAT-------GCCAT----------------------------AAATTTGCC------------------------------------------------------------------------------------CCAA----------------------------------AA--------CGGA------AACTC--------GT--------------------------------------------------TGACA---------------------A----C--CTCTCTC----TCTC------TA--TCTCT--CTT--TCTCT--CT------TTCTCTCTCT----CTCTCTC--T--CTC--TCTCTCT----CTCTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6500:25676277-25676503 + |
|
AATAAGAACC---------------------------------------------------------------------------------GA-TGATTGGGCATG-AGCACAACAC----------------------------TAATTTGTA------------------------------------------------------------------------------------TC-------------------------------------A--------TCTACTATAAATA-------AATGC--------------------------------------------------ATGCA---------AGCCTGGCAAGCC----A--CTCT--CTCTCCCTC----------CCTCT--CTCTCT--CT--CT--CTCTCTCTCT----CTCTCTC------T--CGCTCT--CTCTCTGGCTC--------------------------------------------CGTCT--------AT------C-------------------GGTGCTT-----CTGTGTGTGGCTCGTGT-------------------------------GTGCTGTCACATGGTCGCACTGACTGACCCACTTGGCTGGCTGTCTGGC------------------------- |
| droGri2 |
scaffold_15203:3157469-3157639 - |
|
GTTGGCAAAT--------------------------------------------------------------------------GGAAAATTGTGACTTTGTAATG-C-GCCT-------------------------------------------------------------------------------------------------------------------------------------------------------AAGTACTTGGAAAATAATTCAATTTAATA-------TACTTAAT-------------------------------------------------------------------TATGTACTA------TCATTCTCTGTT----TCCC----------TTCCC--CTCTAT--CTCTCTCTCTCGCTCTCT--------CTCTC----TCTCTCTCT--C--T--CTTTC----------------------------------------------------TCTCTTT----------------------------------------------------TG--------------------CTGGGGGACAATG------------------------------------------------------------------------- |
| droAna3 |
scaffold_13117:5239325-5239596 - |
|
CACCGAGAAGTGCG-CCGCCGG-CACCACAGCCTGCTACTCCTGCGAGCA-AGGTTAG-G------GTGAGTGGGGGACCTAATTGGAGGCCT-TAATCGGGCAA--G-GAATAAAA----------------------------GAGATTATCCTTTATGAGATAGATGTTTTAATGGAAA---------------GGATAACCTTCAGGATCCTTGACTCTCCTTTAAGTTCTTCTACAA----------------------------------AA--------AATAAGAGAAATAAA--------A-----------------------------------------------------------------------------A----C--CAATTAT----CCCT-------------------------------------------------------------------------------------------------------------------------------------------------------T---------------------------------------------------------------AACACAGATACCTTCTTGCGACGTGGCTGTGCGGATG--CGAACTT--TACTCCATCGG--------------------------- |
| droBip1 |
scf7180000395805:34267-34387 + |
|
CTACGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT--C----TCTT------TCTCTCTCTTTGTCGCT------CT--CTCTCTCTCTCCGT----CTCTTTATC----T------CTCTCTT------ACT--------------------------------------CTATTTATATGTC-----------------------------TTTCTCT-----A-------------TG--------------------AGTCCAGATAACCC-----------------------------CTAACCTGAAGCTTAGAT------------------------- |
| droKik1 |
scf7180000302724:209241-209369 - |
|
CTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--CTCTCTCTC----------TCTCT--CTCTCT--CT--CT--CTCTCTCTCT----CTCTCTC------T--CTCTCT--CTCTCTCTCTC--TT-------------------------------------------------CTCTTT------C--TTTGCTCTGCCAACAAACTTCTTT-----T----TCCGGCCTTTGCGCATGAGTCA---------------------ATG--GCTGC-------------------------------------TT------------------------- |
| droFic1 |
scf7180000454072:680971-681263 + |
|
CACGGAGAAGTGCG-CCCCGGG-CACTGCGGCCTGCTACTCCTGCGAGCA-AGGTTAG-GCTCCGAGCCAGTGGAGG------------------------------------------------------------------CT---------CTTTTCCAAATAAAACGTGGT------TGCATCGATCGATAAACCGTCGCTTGCGAGA------------------------------------------------GTT----------------------------------------------TCAATCGCCAAACCAAATCGGGGGG------------------TGC------AATCACCGCACTGATAT---------------------GTA----CTTG----------TACTC--CTAGTA--ATCCTCCTTTCCAAGTGC--------CCT--TG--TAACCACCA--A--C--TTCTT----------------------------------------------------CCC--------------T---------------------------------------------------------------TGTGCAGACACCTTCCTGCGACGGGGCTGTGCGGACG--CGGACTT--TGCTCCTGCTGCT------------------------- |
| droEle1 |
scf7180000491272:1296939-1297058 + |
|
TGCAATAAT--------------------------------------------------------------------------------------------------------------------------------------------TTATC------------------------------------------------------------------------------------AGAA----------------------------------AA--------TATG--------------------T--------------------------------------------------GTCCA---------TT-------------GCAGTCTCTCTC----TCTC----------TCTCT--CTCTCT--CTCTCTCTCTCTCTGTCT--------CTC--TG--TCTCTCTCT--C--T--CTCTA----------------------------------------------------TCTCTGTGGCTATGTC-----------------CTTCTC-------------------------------------------------------------------------------------------------------------------------------- |
| droRho1 |
scf7180000762169:12304-12408 - |
|
CTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--CTCTCTCTC----------TCTCT--CTCTCT--CT--CT--CTCTTTCTCT----CTCTCTC------T--CTCTCT--GTCTCTGTCTC--GGT------------------------------------------------CT--GT-----------------------------------------------------------------------------------------------------------------------------CCCATTCGCTTTAGATACCCTGCAGGATGTGAGTG |
| droBia1 |
scf7180000301760:4113829-4113956 - |
|
TGTTCG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCT------CTCCC----TCTC--TCTCT--CTC--T------CT--CTCTCTCTCTCTCT----CTCTCTCTC----TC--TCTCTCG----ATCCAATG--------------------------------------CTAGTG------C-----------------------------CTAACTT-----T-------------TGTGCATAGAGCA---------------------AGACGGCTGCGTCGCAGCGCTGCCG--CCGGCT----------------------------------------- |
| droTak1 |
scf7180000415522:8795-8870 - |
|
ATTC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--CTCTCTCTC----------TCTCT--CTCTCT--CT--CT--CTCTCTCTCT----CTCTCTCTGTCTC----T------CTCTCTCCCTC--C-------------------------------------------------TCTCTCT-----------------------------------------------------------------------------------------------------------------------------------CTCT------------------------- |
| droEug1 |
scf7180000409061:69510-69814 - |
|
AACGGAAAAGTGCA-CAGCGGG-AACCACAGCCTGCTATTCCTGCGAGCA-AGGTTAG-G------GTGAGTGGGTGGACCTTTCGG------------GGGAATCAG-GGCTACC-TA--TACCCAACAAA-G---------TA---------CATTTATAAATAAAATTGATT---------------------------------------------------------------TCAAAAAGAAGCGTTGTGAAGAGTTGTAGTCCTTTGA------------------------------------AAACCAAAATCCAAGTCA------TTTTCTGATTGATATTC--------------------------------------------------------------------------------------------------------------------------------------------AAATCCTATTAAGATTGTTAAACATTTAAGACACCT-----------------------------------------------------------------------------T----------TTTCGAATTACTGCAGATACCTTCCTGCGGCGAGGCTGTGCGGACA--CGGACTT--TGCACCTGCCGCT------------------------- |
| dm3 |
chrX:5627698-5628032 + |
|
AACGGAAAAGTGCG-CAGCAGG-CACCACGGCCTGCTATTCCTGCGAGCA-AGGTTAG-G------GTGAGTGGGGGAA----T-------CTCGACC-AGGAAC----GGCCCCT-TAAGTACCCACTCAAACTGTCAGTTATT---------CATTTGGAAATAAAATTTGTTAAGCAAT---------------GCATCTCTTCCGGGA------------------------------------------------ATT----------------------------------------------TCAGTTG--------AATTGG----CTTTCATTCTTGG-----TTA------AGTTTCTTAATTGATAA-----------------------------------------------------------------CTACTTAGTTTCAT----TTTTTTAC---------------------GG----A----------TA-TCAACGCTTGAGATACCCTTGTAACCCATTT---CCC-----------------------------------------------------------------------TTTTATAATCCCAAATACCTTTCTGCGACGTGGCTGTGCGGATG--CGGATTT--TGCACCCGCTGCT------------------------- |
| droSim2 |
x:5855051-5855144 - |
|
TGCAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTG------CTCTC----TTGC--TCCCT--CTC--TCTCT--CT------CTCTCTCTCT----CTTTATCTG----TC--TCTCTCC----CTCTGCTG--------------------------------------ACATTT------A-----------------------------AATGTTT-----T-------------CG-------------------------------------------------------------------------CGTGCAAA------------------------- |
| droSec2 |
scaffold_0:16131756-16131880 - |
|
CTG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--CTCCCTCCC----------TCTCT--CCG--TCTCT--CT--CTCTCTCTCT--------CTCTCTC--T--CTCTCT--CTCT----CTCTAGTT--------------------------------------TTGCCT--------CA------------------------------CTTTTCTGGCCTTTCAGCTTCGTGT----------TTTCGTATATTGGCAAACA------------------------------------------CTTTTCGGC------------------------- |
| droYak3 |
X:3423428-3423535 - |
|
AT-----------------------------------------------------------------------------------------------------------------------------------------------AAACGTGGT------------------------------------------------------------------------------------CCAA----------------------------------AG--------T--------------------------------------------------------------------------------------------TAATGAGCAATTCA--CTCT--CTCTCTCTC----------TCTCT--CTCTCT--CT--CT--CTCTCTCTCT----CTCTCTC------T--CTCTCT--CTCTCTCTCTC----------------------------------------------TCT----CTCTCT-----------------------------------------------------------------------------------------------------------------------------------CTTT------------------------- |
| droEre2 |
scaffold_4770:5447746-5447855 + |
|
TATACGAATGTATA-TCAATG---------------------------------------------------------------------------------------------------------------------------------TAAA------------------------------------------------------------------------------------CCAA----------------------------------AG-------------------------------------------------------------------------------------------------------CTG------CCGTGCTATCTC----TTTC----------TACCT--CTCTCT--CTCTCTCTCTCTCTCTCT--------CTCTC----TCTCTCTCT--C--T--TTCTC----------------------------------------------------TATCTGT-----------------------------------------------------------------------------------------------------------------------------------CACT------------------------- |