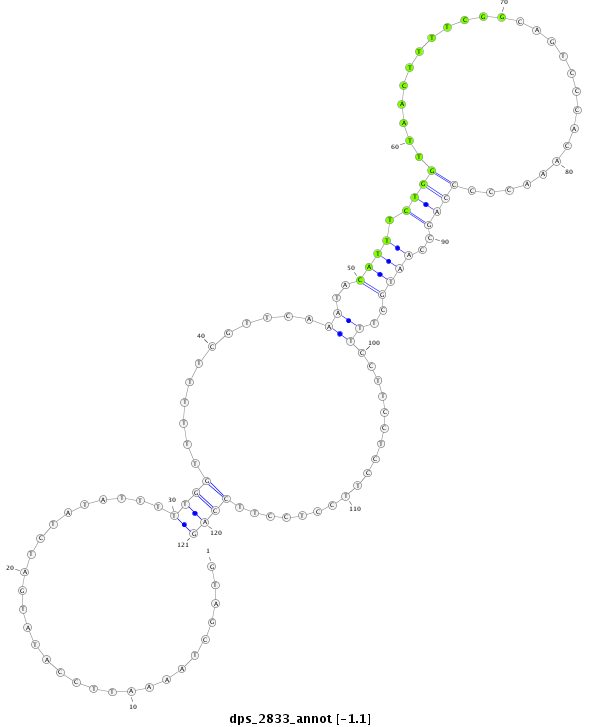
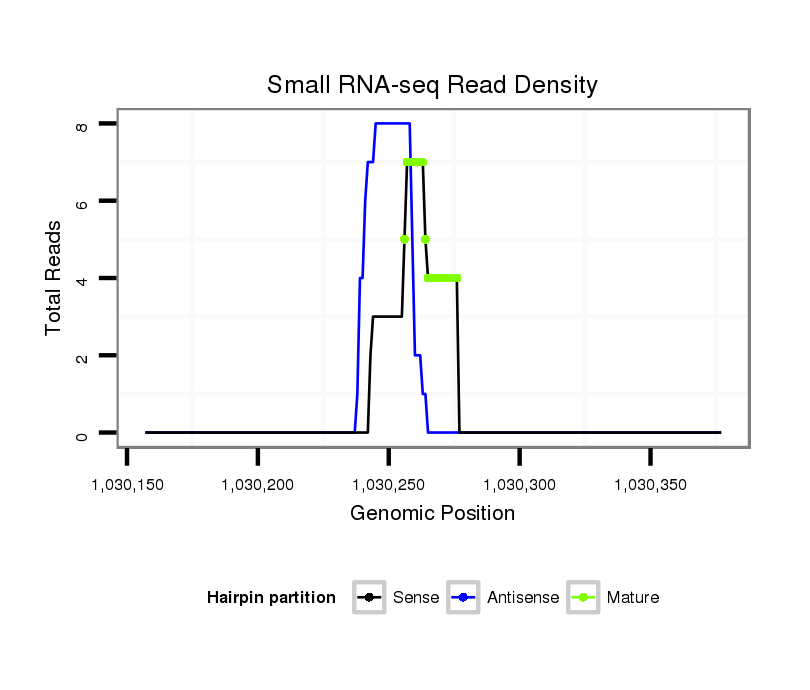
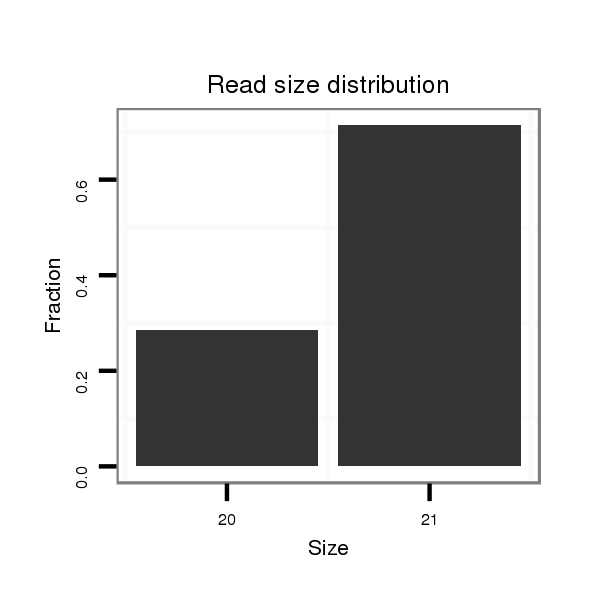
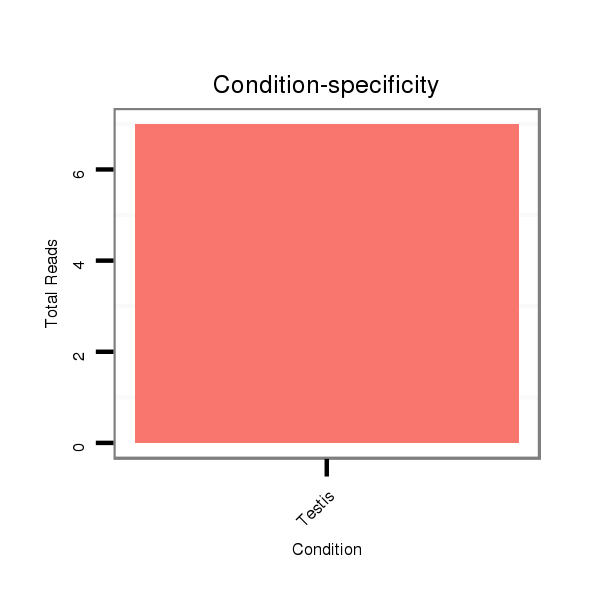
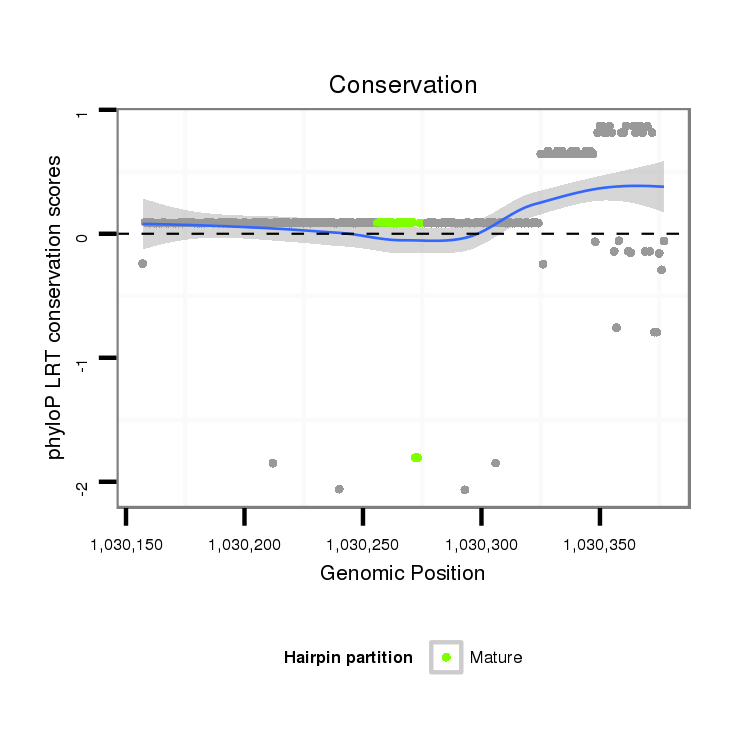
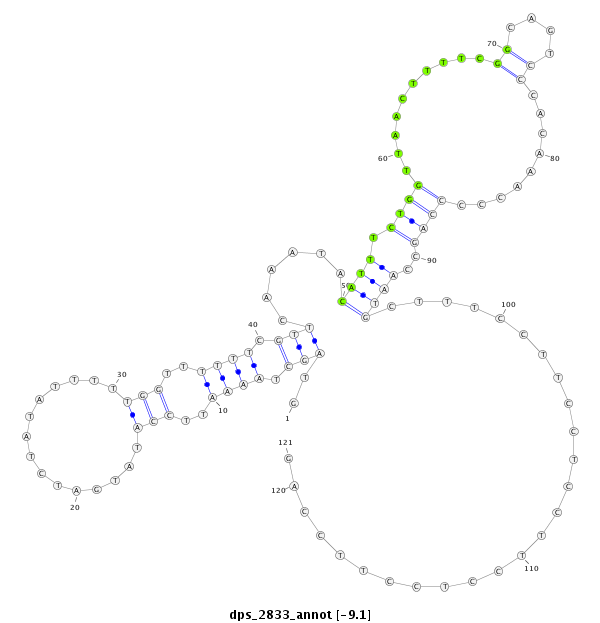
ID:dps_2833 |
Coordinate:XL_group3a:1030207-1030327 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
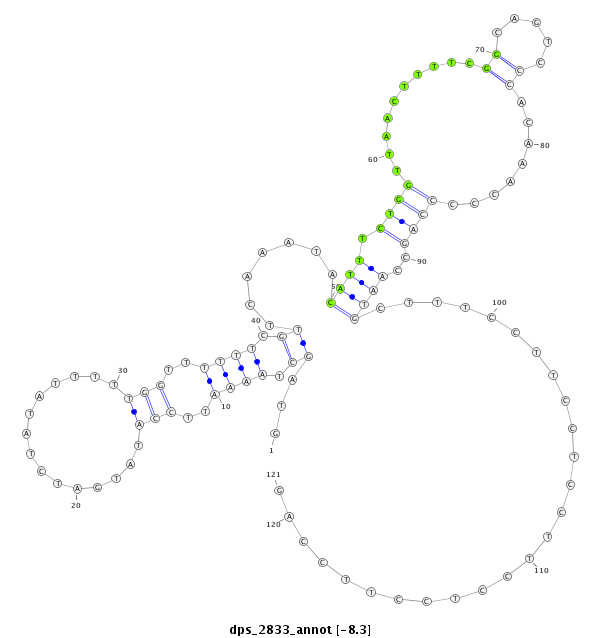
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.1 | -8.7 | -8.7 | -8.3 |
 |
 |
 |
 |
exon [dpse_GLEANR_10313:4]; CDS [Dpse\GA22360-cds]; exon [dpse_GLEANR_10313:3]; CDS [Dpse\GA22360-cds]; intron [Dpse\GA22360-in]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------################################################## AAAGAGCCCTCGGCGGAGAACTCGCGCTCACGCTCTCACTCGCAGCCATAGTAGCTAAAATTCCATATGATCTATATTTTTGGTTTTTTCGTTCAAATACATTTCTGGTTAACTTTTCGGCAGTCCCACAAACCCCCAGCCAATGCTTTCCTTCCTCCTTCCTCCTTCCAGTGGCAGTGGCAGTGGCAGTGGCTTCCAGGGACCAACTGTTGCTGCTTGGC **************************************************.............................((((............((..((((.((((...........................))))..))))..))..................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|---|
| ...................................................................................................CATTTCTGGTTAACTTTTCGG..................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ....................................................................................................ATTTCTGGTTAACTTTTCGG..................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ......................................................................................TTTCGTTCAAATACATTTCTG.................................................................................................................. | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ..............................................................................................................................CACAAACGCCCAGCCAATG............................................................................ | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .......................................................................................TTCGTTCAAATACATTTCTGG................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................................................GGCAGTGGCAGTGGCTTCC........................ | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................AGTCGCTTCCCGGGACCA................ | 18 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ...................................................................................................................................................................................CCAGTGGAAGTGGCTTCCAG...................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 |
| ........CTCGGCCGGGAAGTCGCGC.................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
|
TTTCTCGGGAGCCGCCTCTTGAGCGCGAGTGCGAGAGTGAGCGTCGGTATCATCGATTTTAAGGTATACTAGATATAAAAACCAAAAAAGCAAGTTTATGTAAAGACCAATTGAAAAGCCGTCAGGGTGTTTGGGGGTCGGTTACGAAAGGAAGGAGGAAGGAGGAAGGTCACCGTCACCGTCACCGTCACCGAAGGTCCCTGGTTGACAACGACGAACCG
**************************************************.............................((((............((..((((.((((...........................))))..))))..))..................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
SRR902011 testis |
V112 male body |
GSM343916 embryo |
SRR902012 CNS imaginal disc |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................GTGTTTGCGGGTCGGTTACGT.......................................................................... | 21 | 2 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................AAAAAGCAAGTTTATGTAA...................................................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................CAAAAAAGCAAGTTTATGTA....................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................AAAAAAAGCAAGTTTATGTA....................................................................................................................... | 20 | 1 | 2 | 1.50 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TTGTGTTTGCGGGTCGGTTACG........................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................AGCAAGTTTATGTAAAGACC................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................AAAAGCAAGTTTATGTAAAGA................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................GTTTGCGGGTCGGTTACGT.......................................................................... | 19 | 2 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CAAAAAAGCAAGTTTATGTAA...................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................CCAAAAAAGCAAGTTTATGTA....................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................TGTGTTTGCGGGTCGGTTACG........................................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TCGGTATCAACAATTTTAAAGT............................................................................................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................AAGGAAGGAGGAAGGAGGGA...................................................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................TGTGTTTGCGGGTCGGTTACGT.......................................................................... | 22 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AAGGAAGGAGGAAGGAGAA....................................................... | 19 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................TCAGGGTGTTGGGGG..................................................................................... | 15 | 1 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 3 | 0 |
| .....................................................................................................................................................GAAAGGAGGAAGGAGGAAG..................................................... | 19 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CCTGGGTGACAGCGACGAAGC. | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................AAAGCCGTCGGGATGCTTG........................................................................................ | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................GAAAGGAGGGAGGTAGGAG......................................................... | 19 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................GAAAGGAAGGGGGAAGAAG......................................................... | 19 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................TTTCTGGATCGGTTACGAA......................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................AGACTAAGCGTCGGTATTA......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................GGAGTAGGAAAGAGGAAGGT................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group3a:1030157-1030377 + | dps_2833 | AAAGAGCCCTCGGCGGAGAACTCGCGCTCACGCTCTCACTCGCAGCCATAGTAGCTAAAATTCCATATGATCTATATTTTTGGTTTTTTCGTTCAAATACATTTCTGGTTAACTTTTCGGCAGTCCCACAAACCCCCAGCCAATGCTTTCCTTCCTCCTTCCTCCTTCCAGTGGCAGTGGCAGTGGCAGTGGCTTCCAGGGACCAACTGTTGCTGCTTGGC----- |
| droPer2 | scaffold_15:1863032-1863249 - | AAAGAGCCCTCGGCGGAGAACTCGCGCTCACGCTCTCACTCGCAGCCATAGTAGCAAAAATTCCATATGATCTATATTTTTGGGTTTTTCGTTCAAATACATTTCTGGTTAACTTCCCGGCAGTCCCACAAACCCCAAGCCAATGCTTTTCTTCCTCCTTCCTCCTTCCAGTGGCAGTGGCAGTGGCAGTGGCTTCCAGGGACCAACTGTTGCTGCTT-------- | |
| droAna3 | scaffold_13047:302761-302784 - | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGTGGCAGTGGCAGTGGCAGTGG---------------------------------- | |
| droTak1 | scf7180000415872:452277-452307 + | G----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTTCCAGTTTCCACCTGTTGATTCCCTCT----- | |
| droYak3 | X:18389446-18389479 + | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTCCAGTATCCACTTGTTGATTCAATCTTTCGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/18/2015 at 05:32 AM