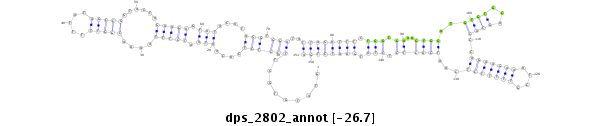
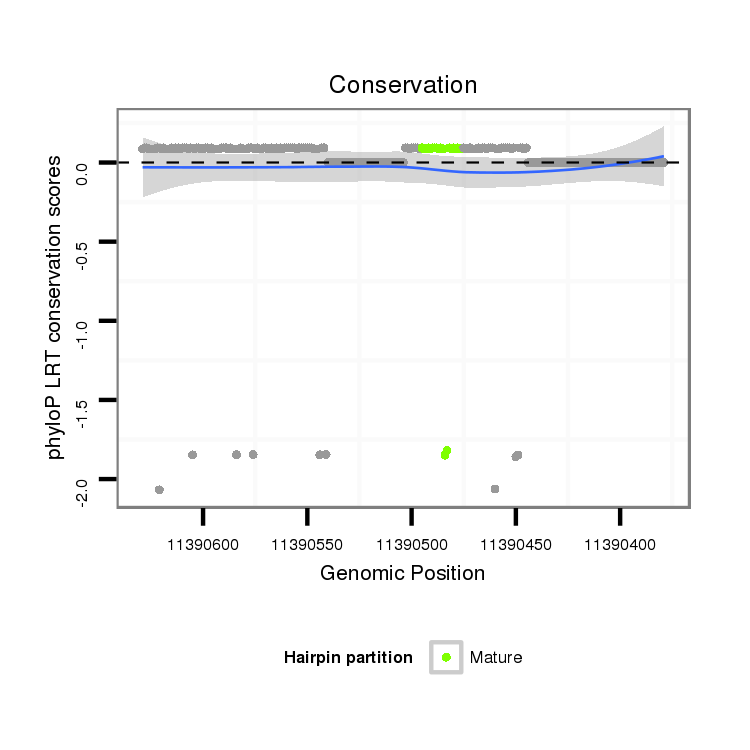
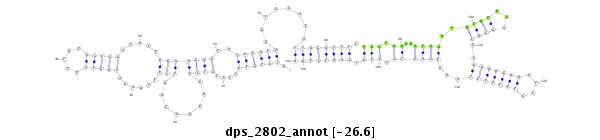
ID:dps_2802 |
Coordinate:XL_group1e:11390429-11390579 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -26.6 | -26.5 | -26.3 |
 |
 |
 |
exon [dpse_GLEANR_74:4]; CDS [Dpse\GA28060-cds]; intron [Dpse\GA28060-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GAAGCGGAGCAGCGACTACTTTCTACTTGGATCACCCGATAGTGGTGAGGGTATGTGCTTCCTCTCACAATAATCTTTAAAACGGTTTTTATAGCCGGTACTAGAAGAGTATACATAGGTGGATATTAGATTTCTGGTGAAAGTGGATGTGTGCAGCACCCAGAAGAAATCGTTTTTCTCAATCACTCTCTATGAATCTGATTAAAAATCGAGTATTTTTGTATGCACTTAAACATTAACCTGAATTCTTT **************************************************.........((((((....(((.(((((....(((((.....)))))......))))).)))....))).)))..((((((((.(((.((..((((..((((...))))..(((((((....)))))))...)))))).))).))))))))************************************************** |
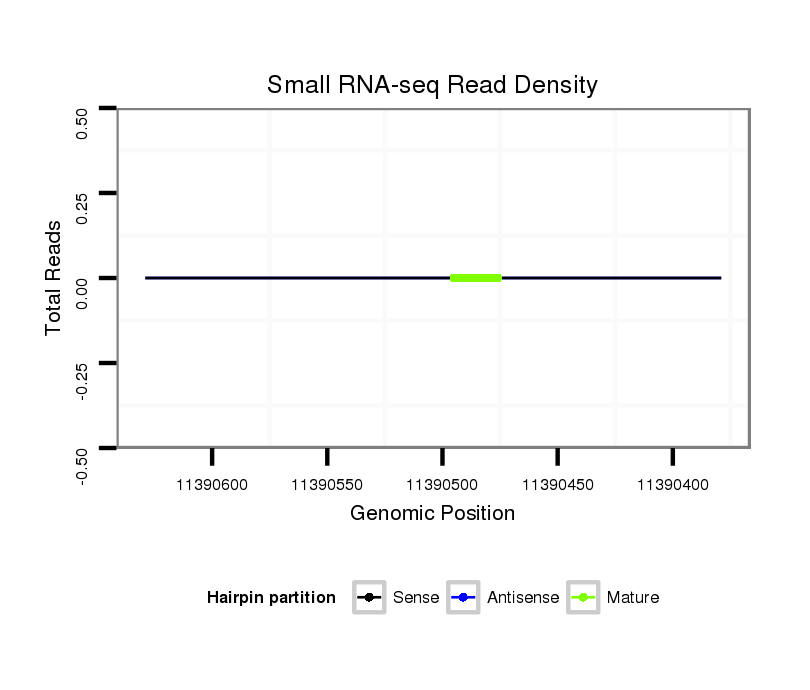
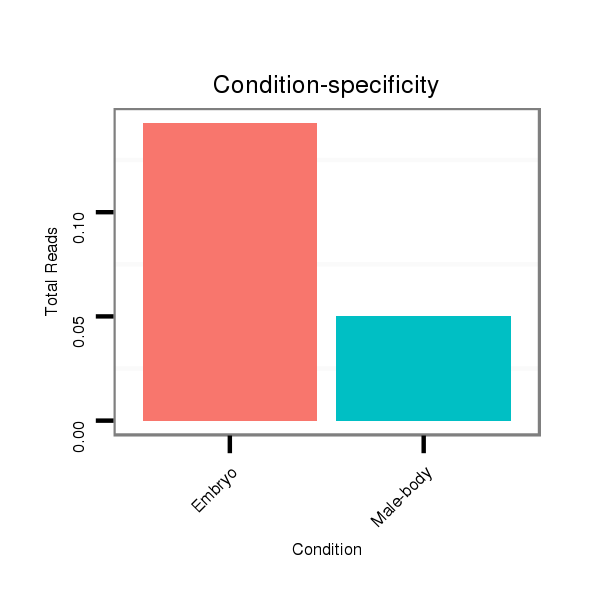
Read size | # Mismatch | Hit Count | Total Norm | Total | M059 embryo |
M040 female body |
GSM343916 embryo |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|
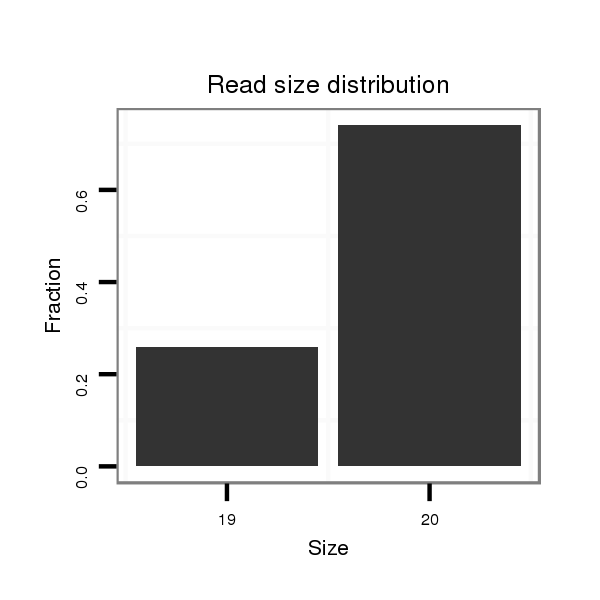
| ......................................................................................................................................TGGTGAAAGTGGATGTGTGC................................................................................................. | 20 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................TATTTTTGTATGTACTTACA.................. | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................TGGTGAAAGTAGATGTGTGC................................................................................................. | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................CTTTAAAATAGTTGTTATAG............................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................TGGTGAAAGTGGATGTGTG.................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
CTTCGCCTCGTCGCTGATGAAAGATGAACCTAGTGGGCTATCACCACTCCCATACACGAAGGAGAGTGTTATTAGAAATTTTGCCAAAAATATCGGCCATGATCTTCTCATATGTATCCACCTATAATCTAAAGACCACTTTCACCTACACACGTCGTGGGTCTTCTTTAGCAAAAAGAGTTAGTGAGAGATACTTAGACTAATTTTTAGCTCATAAAAACATACGTGAATTTGTAATTGGACTTAAGAAA
**************************************************.........((((((....(((.(((((....(((((.....)))))......))))).)))....))).)))..((((((((.(((.((..((((..((((...))))..(((((((....)))))))...)))))).))).))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
GSM343916 embryo |
|---|---|---|---|---|---|---|---|
| ......................GGTGAACCTCGTGGGCTA................................................................................................................................................................................................................... | 18 | 2 | 14 | 0.21 | 3 | 3 | 0 |
| ..........CCGCTCATGAAAGCTGAACC............................................................................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 |
| ..........TTGCTGAAGAAAGCTGAACC............................................................................................................................................................................................................................. | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 |
| .............................................................................................................................................................................AAAAGGGTTAGTGAGAGC............................................................ | 18 | 2 | 13 | 0.08 | 1 | 0 | 1 |
| .......................................................................................................................................................................TTAGCAATAAGAGTTAGTTG................................................................ | 20 | 3 | 15 | 0.07 | 1 | 1 | 0 |
| ........................................................................TAGCATTTTTACCAAAAATAT.............................................................................................................................................................. | 21 | 3 | 15 | 0.07 | 1 | 0 | 1 |
| ................CTGAAAGAACAACCTAGTG........................................................................................................................................................................................................................ | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 |
| ..........................................................................GCAGTTTTGCCGAAAATAT.............................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1e:11390379-11390629 - | dps_2802 | GAAGCGGAGCAGCGACTACTTTCTACTTGGATCACCCGATAGTGGTGAGGGTATGTGCTTCCTCTCACAATAATCTTTAAAACGGTTTTTATAGCCGGTACTAGAAGAGTATACATAGGTGGATATTAGATTTCTGGTGAAAGTGGATGTGTGCAGCACCCAGAAGAAATCGTTTTTCTCAATCACTCTCTATGAATCTGATTAAAAATCGAGTATTTTTGTATGCACTTAAACATTAACCTGAATTCTTT |
| droPer2 | scaffold_14:838983-839130 - | GAAGCGGATCAGCGACTACTTTCTGCTTGGATCACCCGATAGTGGCGAGGGTAAGTGCTTCCTCTCACAATAATCTTTAAAACGGCTTA-------------------------------------TAGATTTCTGGTGAAAGTGATTGTGTGCAGCACCCAGAAGAAAGCGTTTTTCTTGATCA------------------------------------------------------------------ |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 05:30 AM