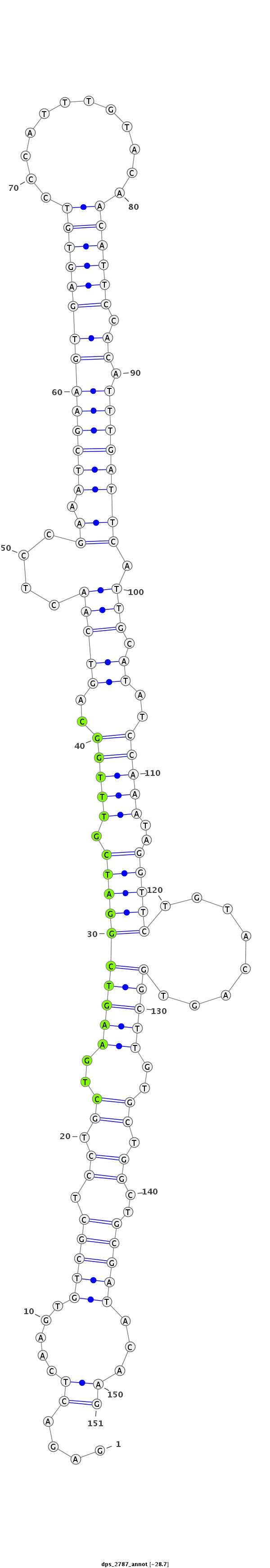
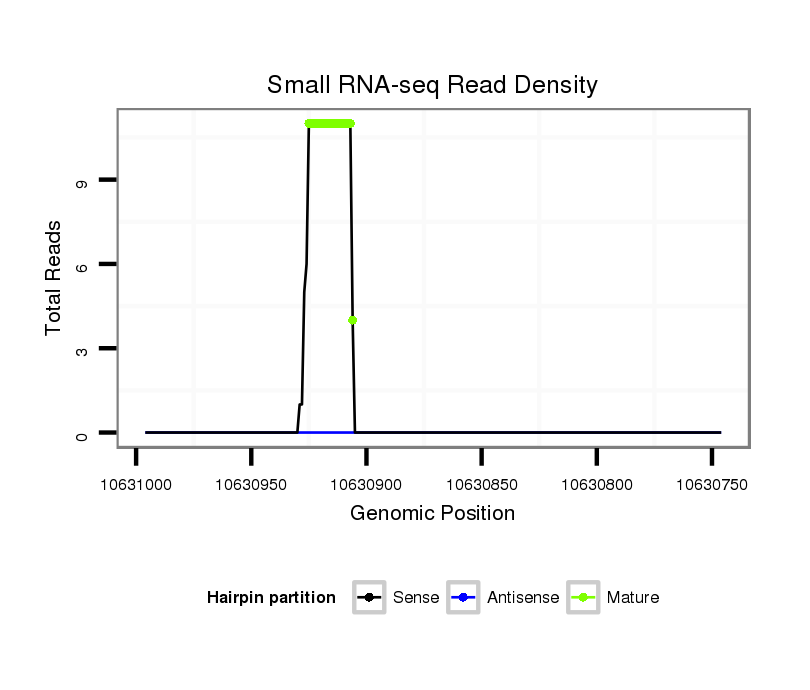
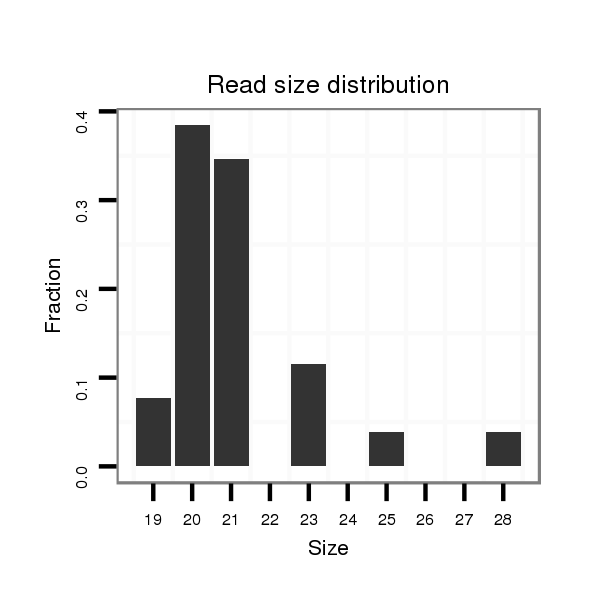
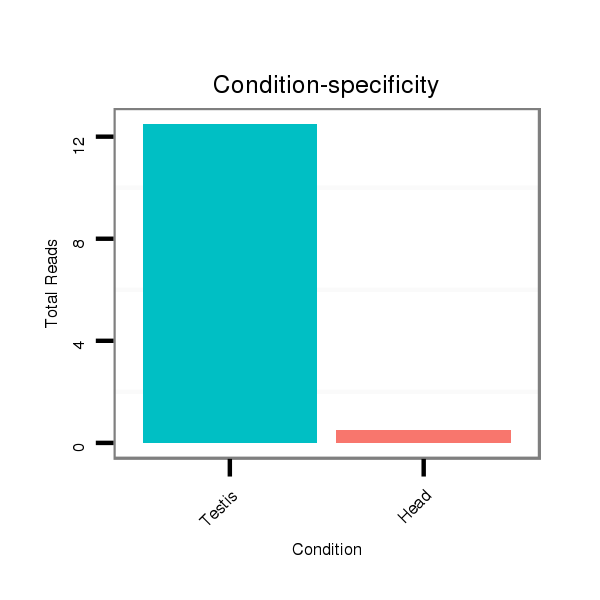
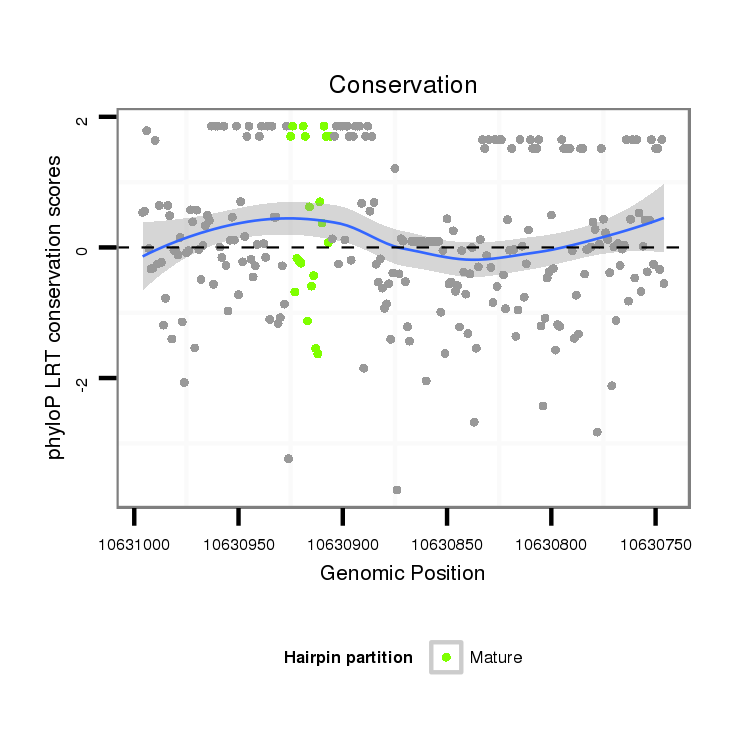
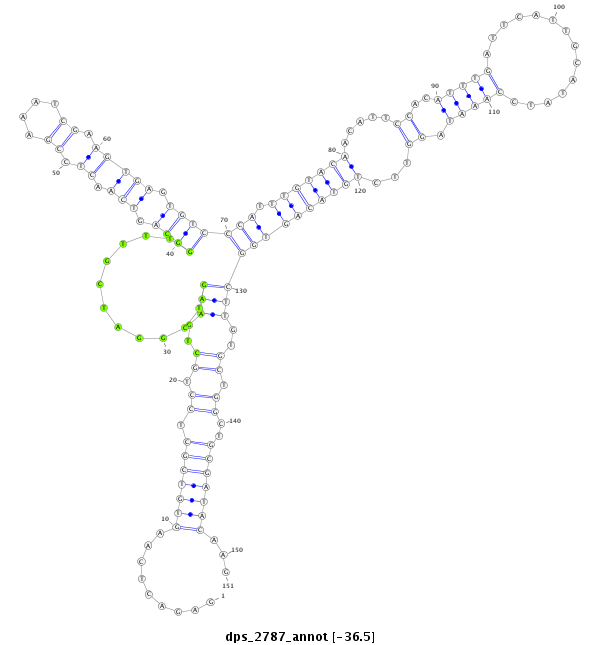
ID:dps_2787 |
Coordinate:XL_group1e:10630796-10630946 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -36.6 | -36.6 | -36.5 | -36.5 |
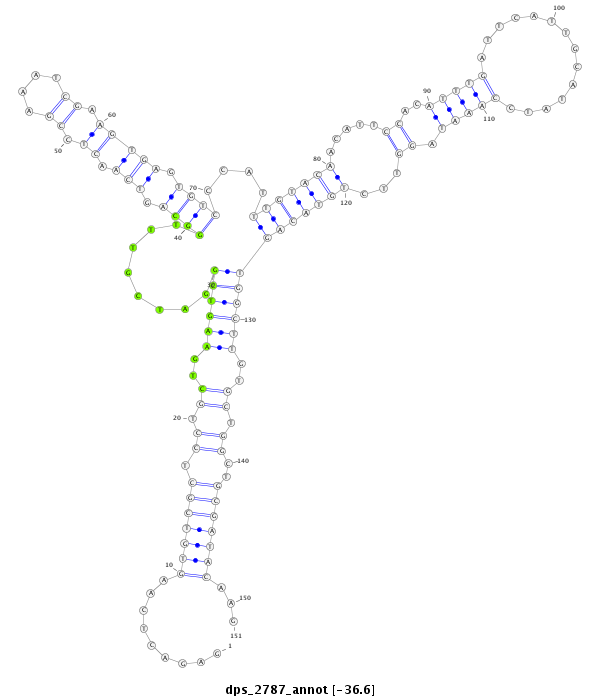 |
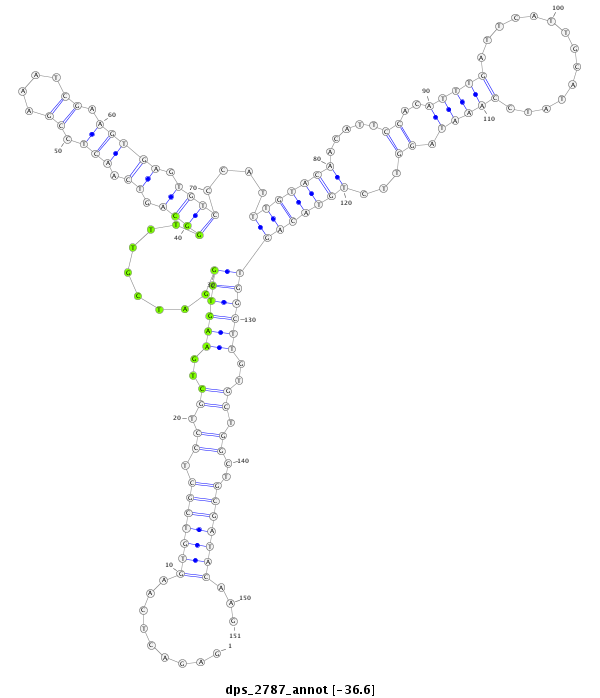 |
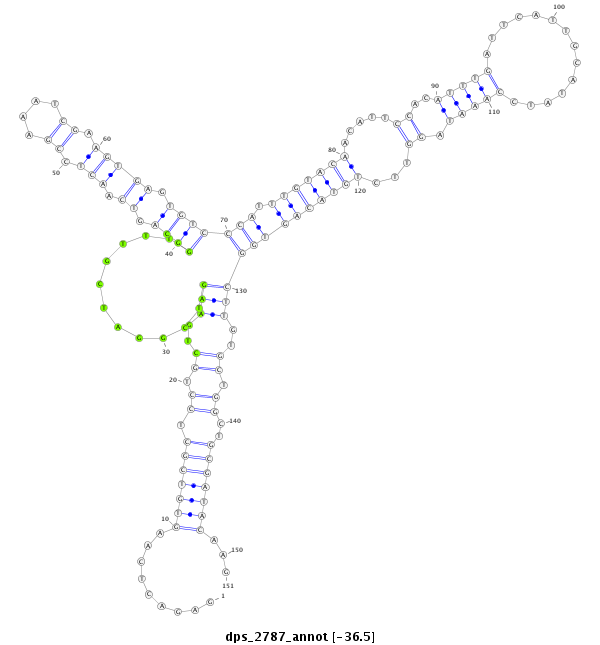 |
 |
exon [dpse_GLEANR_105:2]; CDS [Dpse\GA20339-cds]; intron [Dpse\GA20339-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CCAAGTCCACTCTGGTCATCTTCTCCTCCAAGCAGTTCCTGCGGCTGAGTGAGACTCAAGTGTCGCTCCTGCTGAAGTCGGATCGTTTGGCAGTCAACTCCGAAATCGAAGTGAGTGTCCCATTTGTACAACATTCCACATTTGATTCATTGCATATCCAAATAGGTTCTGTACAGTGGCTTGTGCTGGCTGCGATACAAGTGGCCCATGCGTCGTGGCAGCGTTGGCAAGGTCCTCAATAGCGTCCGCTA **************************************************....((.....(((((.((.((..((((((((((.(((((..(((((....((.((((((((((((((............)))))).)).)))))))).))).))..)))))..)))))........)))))..)).))..)))))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
V112 male body |
M040 female body |
GSM444067 head |
|---|---|---|---|---|---|---|---|---|---|
| .......................................................................CTGAAGTCGGATCGTTTGGC................................................................................................................................................................ | 20 | 0 | 2 | 4.00 | 8 | 7 | 0 | 0 | 1 |
| .....................................................................TGCTGAAGTCGGATCGTTTGG................................................................................................................................................................. | 21 | 0 | 2 | 4.00 | 8 | 8 | 0 | 0 | 0 |
| ...................................................................CCTGCTGAAGTCGGATCGTTTGG................................................................................................................................................................. | 23 | 0 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 |
| ......................................................................GCTGAAGTCGGATCGTTTGG................................................................................................................................................................. | 20 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| .......................................................................CTGAAGTCGGATCGTTTGG................................................................................................................................................................. | 19 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GTGGTAGCGTTGGTAAGGT.................. | 19 | 2 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 |
| ...........................................GCTGAATGAGACTCGAGTGT............................................................................................................................................................................................ | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ........................................................CAAGTGTCGCTCCTGCTGAAGTCGG.......................................................................................................................................................................... | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CAAGTGGCCCATGCGTCGTGG................................. | 21 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................TTGTGCTGGCTGGGATACAA................................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................CGCTCCTGCTGAAGTCGGATCGTTTGGC................................................................................................................................................................ | 28 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................GCTGAAGTCGGATCGTTTGGC................................................................................................................................................................ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................AGGTTCTGCACAGGGGCT...................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................TGGCCCCTGCGTCCCGGCA............................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
GGTTCAGGTGAGACCAGTAGAAGAGGAGGTTCGTCAAGGACGCCGACTCACTCTGAGTTCACAGCGAGGACGACTTCAGCCTAGCAAACCGTCAGTTGAGGCTTTAGCTTCACTCACAGGGTAAACATGTTGTAAGGTGTAAACTAAGTAACGTATAGGTTTATCCAAGACATGTCACCGAACACGACCGACGCTATGTTCACCGGGTACGCAGCACCGTCGCAACCGTTCCAGGAGTTATCGCAGGCGAT
**************************************************....((.....(((((.((.((..((((((((((.(((((..(((((....((.((((((((((((((............)))))).)).)))))))).))).))..)))))..)))))........)))))..)).))..)))))...))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M040 female body |
V112 male body |
GSM343916 embryo |
GSM444067 head |
M022 male body |
M062 head |
SRR902010 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................AGATTCGTCTAGGACTCCG.............................................................................................................................................................................................................. | 19 | 3 | 20 | 9.10 | 182 | 178 | 3 | 0 | 0 | 1 | 0 | 0 |
| .........................GAGATTCGTCTAGGACTCCG.............................................................................................................................................................................................................. | 20 | 3 | 6 | 3.83 | 23 | 11 | 9 | 2 | 1 | 0 | 0 | 0 |
| .........................GAGATTCGTCTAGGACTCC............................................................................................................................................................................................................... | 19 | 3 | 20 | 1.05 | 21 | 17 | 4 | 0 | 0 | 0 | 0 | 0 |
| ......................GAAGAGATTCGTCTAGGAC.................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.90 | 18 | 15 | 0 | 0 | 0 | 3 | 0 | 0 |
| .......................AGGAGATTCGTCTAGGACTCC............................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................GAGGAGATTCGTCTAGGACTC................................................................................................................................................................................................................ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................CCGACGCTCTGTTAACGGGGT........................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................TCGCGGACTACTTCAGCCTA........................................................................................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CTGTCGCAACCGTTGCAG................. | 18 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CGTTCCAGTAGTCATCCCA...... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................CTAGCACGACCGACACTAT...................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1e:10630746-10630996 - | dps_2787 | CCAAGTCCACTCTGGTCATCTTCTCCTCCAAGCAGTTCCTGCGGCTGAGTGAGACTCAAGTGTCGCTCCTGCTGAAGTCGGATCGTTTGGCAGTCAACTCCGAAATCGAAGTGAGT--GTCC---CATTTGTA--------------------------------------------------------------------------------------C--------------------------------------------------------------------------------AACATTCCACATTTGATTCATTGCATATCCAAATAGGTTCTGTACAGTGGCTTGTGCTGGCTGCGATACAAGTGGCCCATGCGTCGTGGCAGCGTTGGCAAGGTCCTCAATAGCGTCCGCTA |
| droPer2 | scaffold_14:64173-64435 - | CCAAGTCCACGCTGGTCATCTTCTCCTCCAAGCAGTTCCTGCGGCTGAGTGAGACTCAAGTGTCGCTTCTTCTGAAGTCGGATCGTTTGGCAGTCAACTCCGAAATTGAAGTGAGT--GTCC---CAG-------AA--------------------------------------------------------------TCGGTTCCCATTTGGA----C--------------------------------------------------------------------------------AACATTCAACATTTCATTGATTGCATATCCAAATAGGTTTTGTACAGTGGCTTGTGCTGGCTGCGATACAAGTGGCCCG-GCGTCGTGGCAGCGTTGGCAAGGTCCTCAATAGCGTCCGCTA | |
| droVir3 | scaffold_12875:8444634-8444885 - | CCAAGTCGACGCTGATCATAATCTGCAGTCAGGAATTTCTGCACCTTAGCGAGATGCAGATATGCGCCCTGCTTAGGTCCAGCTCTTTGTCCGTTAACTCTGAAACTGAGGTTGGC--AATGCTCCAATTAGGATTGCTCCAATCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATTGTTTCTTCCCTTCCGTAGCTACTTTATGGTGTGCTGCACTGGCTGGATGTGGACTGGCCCGCTCGGATGGCGTGTGTGGGTAATGTGCTTAAGAATATACGCTT | |
| droBip1 | scf7180000396441:1238-1353 + | CCAAAGCAACTTTGGTAATACTATCCAGTCGTGAGTTTCTTTGCCTCAATGAAAATCAGGTGTGCCATTTGCTAGCATCCAGCTCTTTGGCAGTAAACTCAGAAATGGAGGTGAGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droFic1 | scf7180000454038:924496-924749 + | CCAAATCATTTACCACAATTTCATCAAGTCGGGAATTTCTGTGCCTCAGTGAGATGCAGATTTGCATGTTGCTATCGTCAAACAGTTTGGCAGTCAACTCGGAAATGGAGGTTGAC--CTTTTTG----------GTCATATATTCA-------------------------------------------------------------------TTGTTATATTT-----------------------------------------------------------------------------------------TATTATATTTTCTTTTTTATAGGTTTTTTATAGCGCTCTGATGTGGCTTCGTCATTTATGGCCAAGCCGCCGTTCGAGCGTATATAAAGTTTTGAGTCAAATTCGTTT | |
| droEle1 | scf7180000490994:163560-163811 + | CTAAATCAATGATGAGAGTGGCCTCAAGCCGGGAATTTTTGGGCCTCAGTGAACGGCAGGTATGCAGCCTACTGCAATCAAATAGCCTGGCAGTCAACTCGGAAATGGAGGTAGTC--CTATTTT----------AACTTATA--------------------------------------------------------------------TACTTATTAAATAC-----------------------------------------------------------------------------------------TA-AATACTGTTTATTATGCAGGTTCTGTACAGCGCATTGTTGTGGCTCAATCATTTATGGCCAAAGCGCAGTTCAAGCACTCATAAGGTTTTAAAAAACATTCGTTT | |
| droRho1 | scf7180000776762:64207-64437 + | ---------------------------------AATTTCTCTGCCTCAGTGAGCGGCAGGTATGCAGCTTACTGAAATCAAATAGTCTGGCAGTCAACTCGGAAATGGAGGTAGGC--TTAT-------------------------------TAAAA------------------------------------ACCTATAAGTACTCGTATGGTTATTGCATGC-----------------------------------------------------------------------------------------TTAAAGAGTCCCTTTTTCACAGGTTTTGTATTGCGCACTGTTGTGGCTCAATCATTTATGGCCAAAGCGCCGTTCATGCACTCAGAAGATATTAAAAAACATTCGCTT | |
| droBia1 | scf7180000302421:2476981-2477307 + | CCAGATCAACCATGGCAGTTCTAGCCAGCCGGGAGTTCATGTGCCTTGCTGAGGTGCAAGTGTGCTGCCTTCTGAAGTCCAGTGGTCTGTCGGTCAACTCGGAAATAGAGGTAGGC--CGTCCTT----------GAGTAACA--------------------------------------------------------------------TGAGTAATCTATACTGAAACATGATTTGTTACCTTTATATTGTACAGAAATTGGACATATCATACATATTCTGTATTGCCTTTTAATGA--------------CAGTATA-TTTTTCCTGGACAGCTCTTATATGGCGCCCTATCGTGGCTTCTATACGGCTGGCCAGCACGCCGTTCTAGCACTAACAGGATCTTGAAAAACATTCGCTA | |
| droTak1 | scf7180000415580:422884-423135 - | CCAGAGCCACTTTGTCCATTATAGCTAGCCGGGAATTCTTGAGCATGAGTGAGGTGCAAGTATGCTGCTTGCTGAAATCAAGTTATCTGGCAGTGAATTCGGAAATTGAGGTAATGTACATACTAAAAAAAGCAAGATTTCAATAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTTATCTTTTCTACTCCAGCTTTTATATAGTGCTTTGATGTGGCTCTTTCATCACTGGCCAGAACGCCAATCCAGCACTGTAAAGGTTTTGAAAAACATTCGTTA | |
| droSim2 | x:14828719-14828992 + | CCACTTCAATTTTGCCAGCCGCATCGACTCGGGAATTCCTGTTCCTCAACGAAATGCAAATTTGCAGCATCCTCAAGTCCAACTTTCTGGCAGTCAATTCGGAAATGGAGGCAAGT--CCTA---TAGCTATATCTATATCTATATATCTGTTTAACT------------------------------------ACCTAGAAGTATAACCGTGATATAT-----------------------------------------------------------------------------------------------------------GTATCCACAGCTTTTATATTCCGCCCTAATGTGGCTAGCGCATCTTTGGCCGGAACGACGTTCGAGCACCTATCCGGTTTTGAATCAAATTCGCTA | |
| droSec2 | scaffold_37:235293-235566 + | CCACTTCAATTTTGCCAGTCGCATCGACTCGGGAATTCCTGTTCCTCAACGAAATGCAAATTTGCAGCATACTCAAGTCCAACTTTCTGGCAGTCAATTCGGAAATAGAGGCAAGT--CCTA---TATCTATATCTATATCTATATATCTGTTTAACT------------------------------------ACCTAGAAGTATAACCGTGATATAT-----------------------------------------------------------------------------------------------------------GTATCCACAGCTTTTATATTCCGCCCTAATGTGGCTAGCGCATCTTTGGCCGGAACGACGTTCGAGCACCTATCCGGTTTTGAATCAAATTCGCTA | |
| droYak3 | X:9855987-9856306 + | ACAGGGCAACTTTGCCAGCCGCATCCACTAGGGAGTTCCTCAGCATCAGCGAGGTGCAAATTTGCTCCATACTGAAGTCCAGTACTCTGGCAGTCAACTCGGAAATGGAGGCAAGT--ACA------TCTATCTGTCAATCTATCTATCTATGTATCTAAATATATGTGGCGAAAGCTTTGATGTTTCCTAATAAGATATAAGTAA--CTATAGTAATCCAATGA-----------------------------------------------------------------------------------------TCCTAA---CTTCAATTCACAGCTCCTATACTGCGCCTTGATGTGGCTATCGCATTGCTGGCCACAGCGCCGTTCCAGCACCAATGTGGTTTTGAAACACATCCGCTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/18/2015 at 05:29 AM