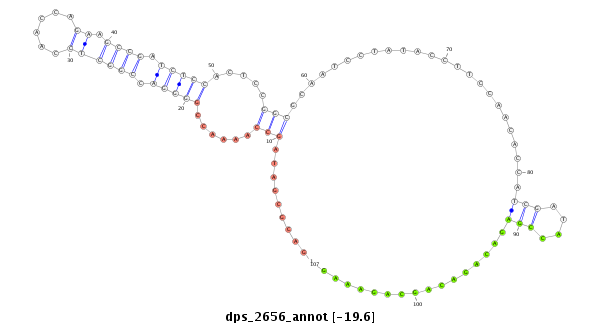
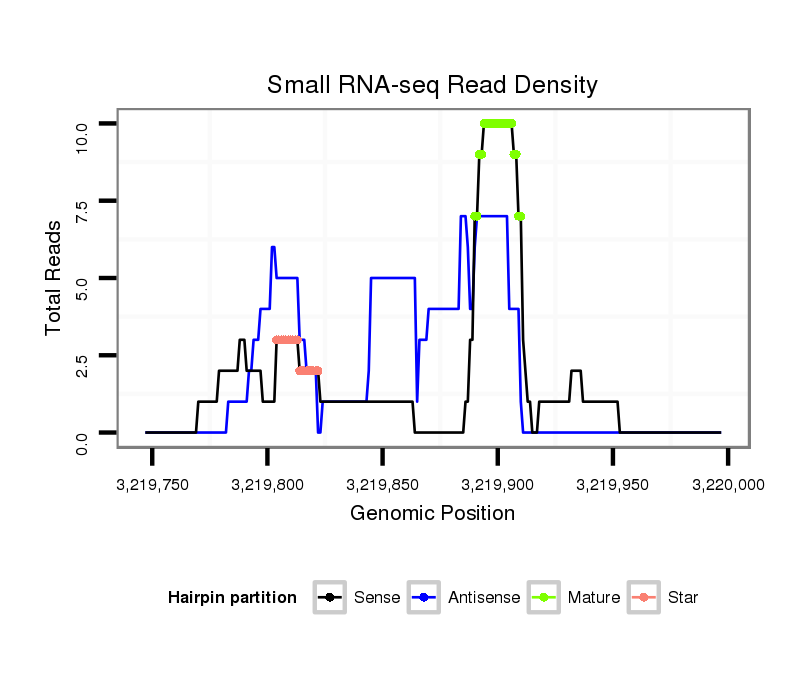
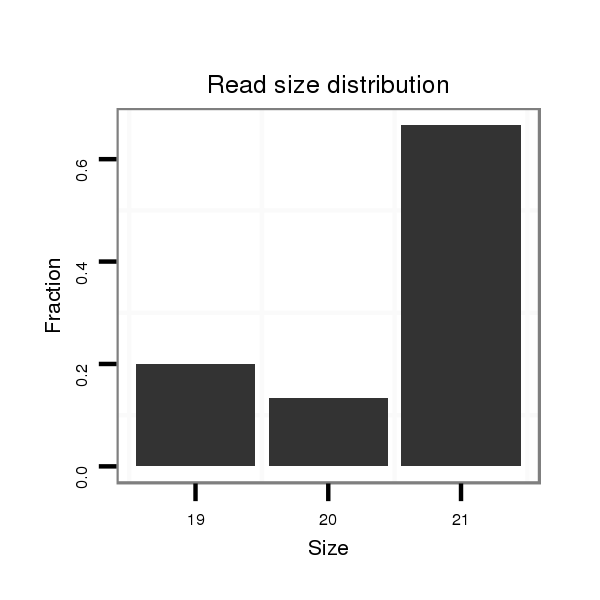
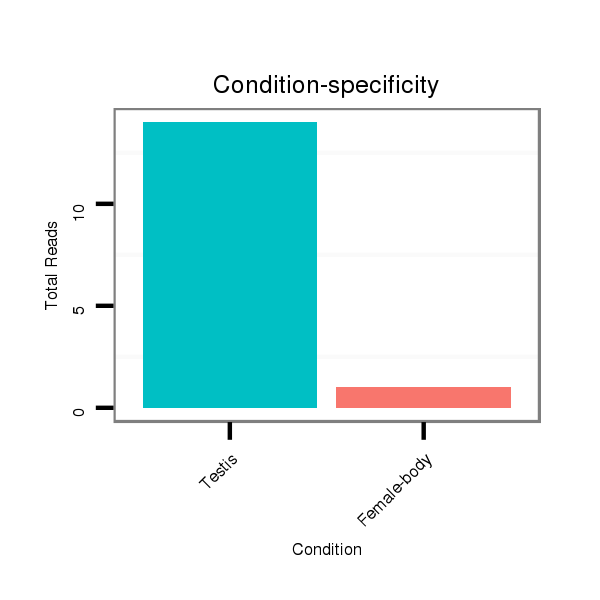
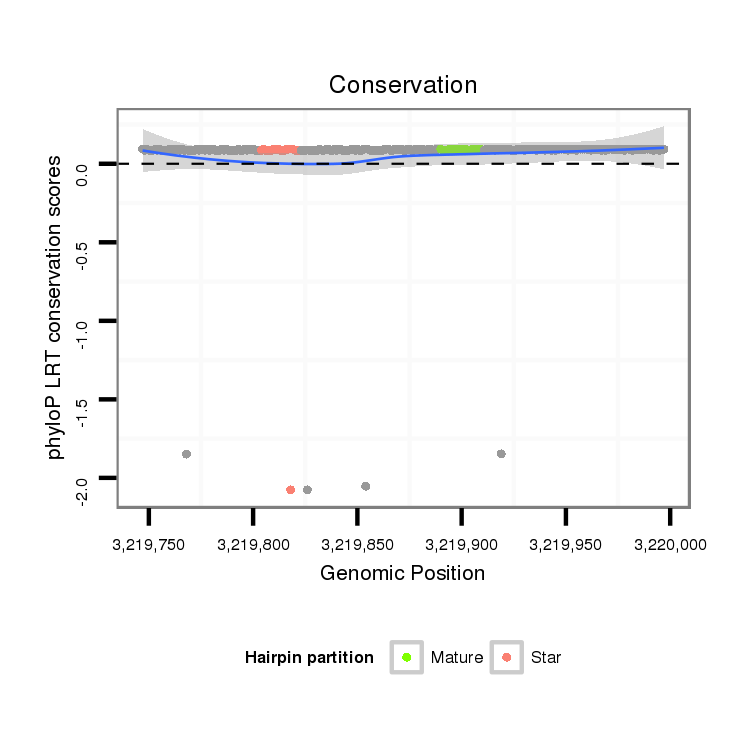
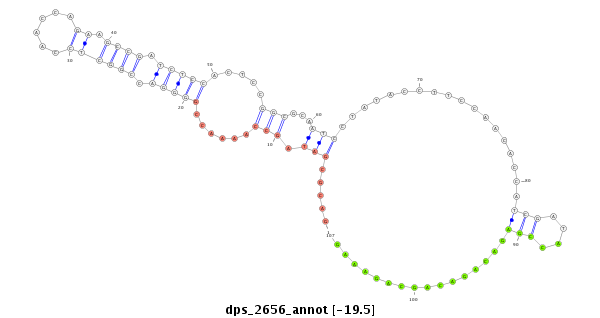
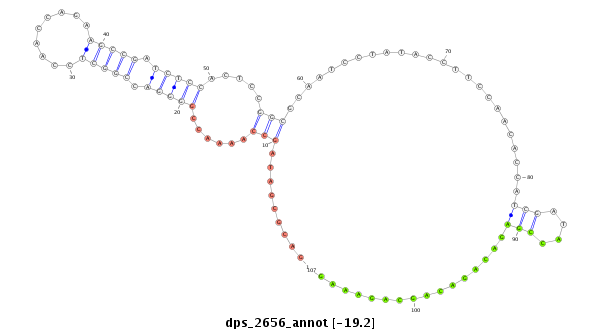
ID:dps_2656 |
Coordinate:XL_group1e:3219797-3219947 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -19.5 | -19.2 | -19.2 |
 |
 |
 |
CDS [Dpse\GA28702-cds]; exon [dpse_GLEANR_881:2]; intron [Dpse\GA28702-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGTCCCTGACCCCTTTATCTGGCCGCAGTAGTCAAGCCAGAGGCCGACCTGTAAGCCGACGCGATAGCCAAAACCGGGGACCGGCTCCAACCAGAAGCCGATCTCCACTCCGGCGCAATCCTATACCTTCCAACACCATCGATACCGAGACAGACAGCAGAAAGGATTTCTTAAAATCGCAATGTAAAACAATGTAGTGATTTTCAAACTAAATTCATTAGACAATATTATCAAACAATATAAAACATGAA *********************************************************.........(((......(((((.((((((......)).)))).))))).....)))........................(((....)))................*************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
GSM444067 head |
M022 male body |
SRR902010 ovaries |
V112 male body |
GSM343916 embryo |
M059 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................ACCGAGACAGACAGCAGAAAG....................................................................................... | 21 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GACGCGATAGCCAAAACCG............................................................................................................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ATACCGAGACAGACAGCAGAA......................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CAAGCCAGAGGCCGACCTG........................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AGACAGACAGCAGAAAGGATT................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................CGAGACAGACAGCAGAAAGG...................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................GGCCGACCTGTAAGCCGACGCGATAG........................................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................GGGACCGGCTCCAACCAGAAG.......................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................CCGATCTCCACTCCGGCGCA...................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CGATACCGAGACAGACAGCAG........................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................AAAACAATGTAGTGATTTTCA............................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TAAAATCGCAATGTAAAAC............................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CGCAGTAGTCAAGCCAGAGGC............................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................CGAGACAGACAGCAGAAAGGA..................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TTCCACCACGACCGATACCG........................................................................................................ | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................AGAGGCCGATCTGTTGGCCGA................................................................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................ATAGCAATGTAGAACAATCT........................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................TGAAAGGATTTCTGAAAATGG........................................................................ | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................GCAGTACTAGTCAAGCCAG................................................................................................................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................CAAAGGCCGATCTGTAAGA................................................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................GAAGACGATCTGCACTCCC........................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TCAGGGACTGGGGAAATAGACCGGCGTCATCAGTTCGGTCTCCGGCTGGACATTCGGCTGCGCTATCGGTTTTGGCCCCTGGCCGAGGTTGGTCTTCGGCTAGAGGTGAGGCCGCGTTAGGATATGGAAGGTTGTGGTAGCTATGGCTCTGTCTGTCGTCTTTCCTAAAGAATTTTAGCGTTACATTTTGTTACATCACTAAAAGTTTGATTTAAGTAATCTGTTATAATAGTTTGTTATATTTTGTACTT
***************************************************************************************.........(((......(((((.((((((......)).)))).))))).....)))........................(((....)))................********************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
V112 male body |
GSM444067 head |
M040 female body |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................TAGCTATGGCTCTGTCTGTCG............................................................................................. | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ...............................................................................................................................................TGGCTCTGTCTGTCGTCTTT........................................................................................ | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| .......................................................................................................................GGATATGGAAGGTTGTGGTAGC.............................................................................................................. | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .......................................................GGCTGCGCTATCGGTTTTGG................................................................................................................................................................................ | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..................................................................................................GCTAGAGGTGAGGCCGCGTT..................................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .................................................................................................GGCTAGAGGTGAGGCCGCGTT..................................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 |
| ...............................................GGACATTCGGCTGCGCTATC........................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................CTGGACATTCGGCTGCGCTATC........................................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................GCTAGAGGTGAGGCCGCGTTA.................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................ATGGAAGGTTGTGGTAGCTA............................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................GGATATGGAAGGTTGTGGTAG............................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................GGTCTCCGGCTGGACATTCGG.................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................CCTGGCCGAGGTTGGTCTTC.......................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..................................................CATTCGGCTGCGCTATCGGT..................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................GGCTCTGTCTGTCGTCTTTC....................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................TTGGTCTTCGGGAAGAGGTG............................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ........TGGGTAAATGGACCGGCGTAAT............................................................................................................................................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................CGACAAGAGGTGAGGCCGC........................................................................................................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 |
| .........................................................CTGCGCGATTGGTTCTGGC............................................................................................................................................................................... | 19 | 3 | 7 | 0.29 | 2 | 0 | 1 | 0 | 1 |
| .........................................................CTGCGCGATTGGTTCTGG................................................................................................................................................................................ | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1e:3219747-3219997 + | dps_2656 | AGTCCCTGACCCCTTTATCTGGCCGCAGTAGTCAAGCCAGAGGCCGACCTGTAAGCCGACGCGATAGCCAAAACCGGGGACCGGCTCCAACCAGAAGCCGATCTCCACTCCGGCGCAATCCTATACCTTCCAACACCATCGATACCGAGACAGACAGCAGAAAGGATTTCTTAAAATCGCAATGTAAAACAATGTAGTGATTTTCAAACTAAATTCATTAGACAATATTATCAAACAATATAAAACATGAA |
| droPer2 | scaffold_17:154839-155089 + | AGTCCCTGACCCCTTTATCTGACCGCAGTAGTCAAGCCAGAGGCCGACCTGTAAGCCGACGCGATAGCCAACACCGGGGCCCGGCTCCAACCAGAAGCCGATCTCCAATCCGGCGCAATCCTATACCTTCCAACACCATCGATACCGAGACAGACAGCAGAAAGGATTTCTTGAAATCGCAATGTAAAACAATGTAGTGATTTTCAAACTAAATTCATTAGACAATATTATCAAACAATATAAAACATGAA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 05:09 AM