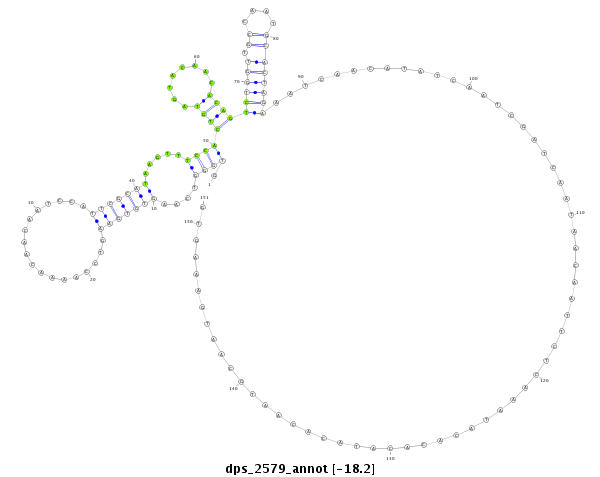
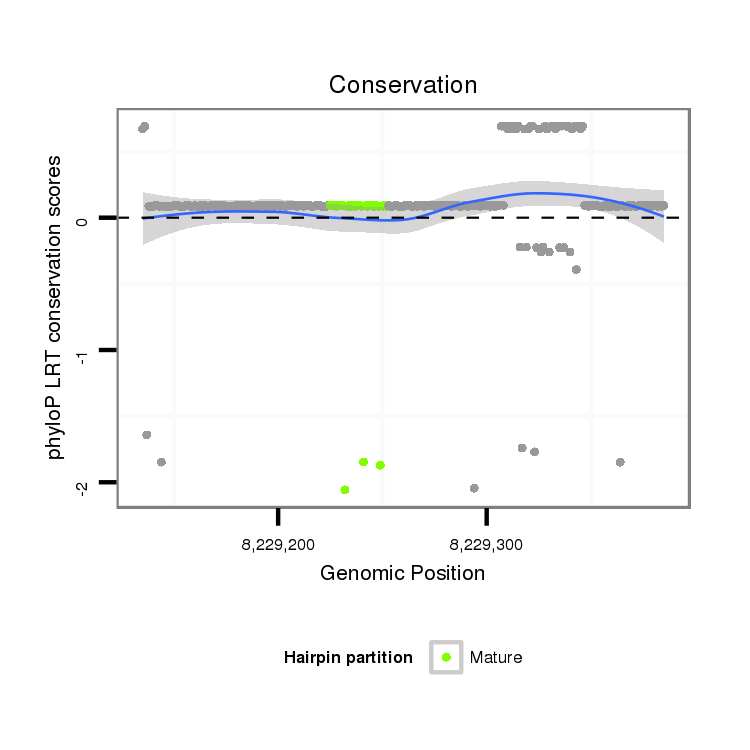
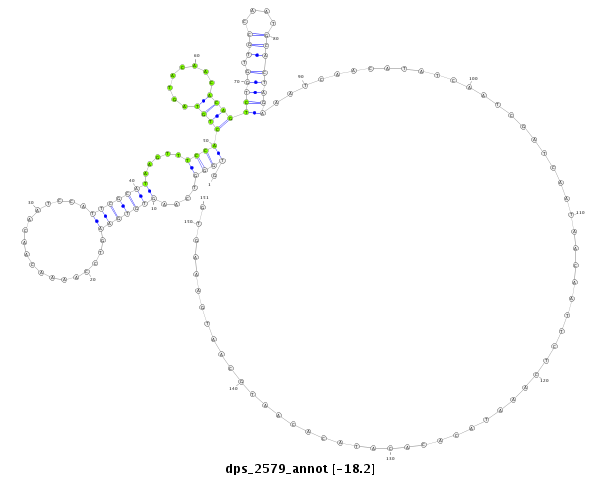
ID:dps_2579 |
Coordinate:XL_group1a:8229185-8229335 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -18.2 | -18.0 | -18.0 |
 |
 |
 |
exon [dpse_GLEANR_11600:1]; CDS [Dpse\GA23089-cds]; intron [Dpse\GA23089-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CATGGCAAGGGCGCCTTTCGCATGGAGGACTGCTTCATAACCGCCACAAAGTGGGTCAAGTGTGAAGTCCAAAACAACAATCCATTCGCATAAGTTTCCACTGTAGTACAACACAGTCTGGTTGCCAATGCACTAGAAATCAACATATCAATCGATCAATAACAATTCTCAAATACACACATACACAATGCAATGAAAGTGTGTAAGTTTGTTTCTTTTCGCTGCGTGCGTCGAGTGGATTGTTGATCAGT **************************************************.((((....(((((((..................))))))).....))))((((........))))((((((.((....))))))))................................................................************************************************** |
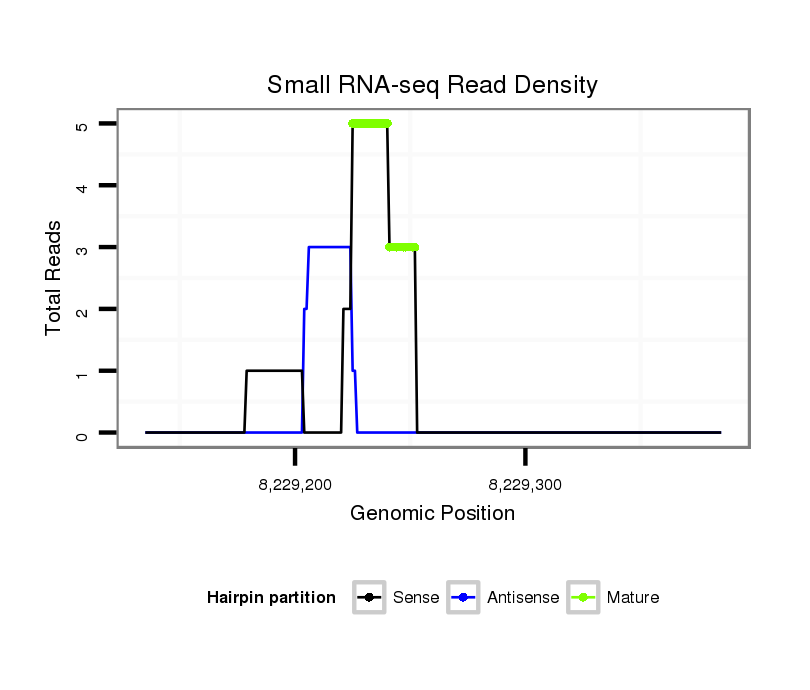
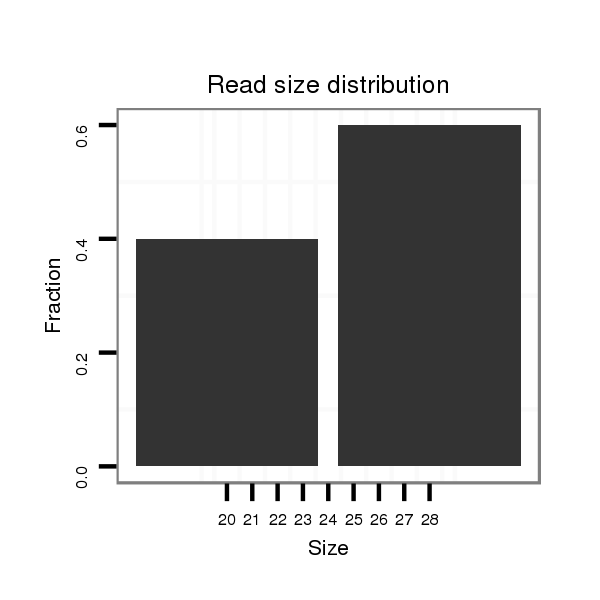
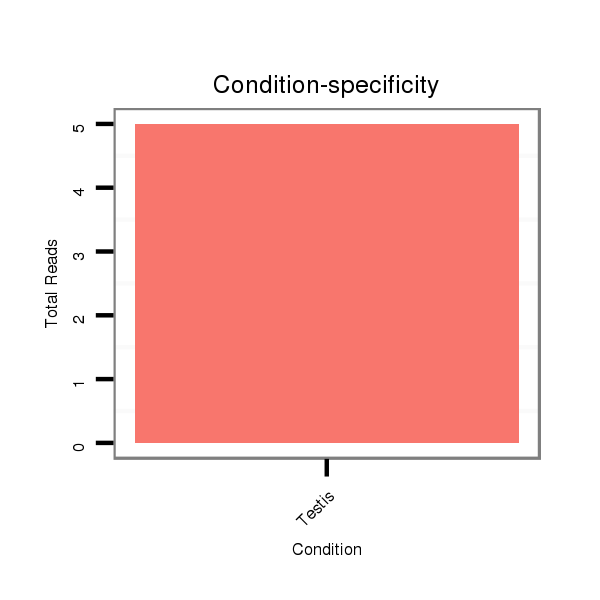
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
GSM444067 head |
GSM343916 embryo |
SRR902012 CNS imaginal disc |
M040 female body |
SRR902010 ovaries |
V112 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................TAAGTTTCCACTGTAGTACAACACAGTC..................................................................................................................................... | 28 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CGCATAAGTTTCCACTGTAG................................................................................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................CACAAAGTGGGTCAAGTGTGAAGTC...................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................AATCATAGTGTGTAAGATTGTT...................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TGCGTGCGTCGAGTGGACTGG........ | 21 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................GCACAATGCACTGAAAGTG.................................................. | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................GCAAGTGTGAGGTCCAAAA................................................................................................................................................................................. | 19 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................................CGGTGGCTTGTTGATCAGT | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................CACAAAGTGGTACAGGTGT............................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................ATGCACAAGAATTCAACA.......................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GTACCGTTCCCGCGGAAAGCGTACCTCCTGACGAAGTATTGGCGGTGTTTCACCCAGTTCACACTTCAGGTTTTGTTGTTAGGTAAGCGTATTCAAAGGTGACATCATGTTGTGTCAGACCAACGGTTACGTGATCTTTAGTTGTATAGTTAGCTAGTTATTGTTAAGAGTTTATGTGTGTATGTGTTACGTTACTTTCACACATTCAAACAAAGAAAAGCGACGCACGCAGCTCACCTAACAACTAGTCA
**************************************************.((((....(((((((..................))))))).....))))((((........))))((((((.((....))))))))................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902011 testis |
M040 female body |
GSM343916 embryo |
SRR902010 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................GTTTTGTTGTTAGGTAAGCGT................................................................................................................................................................. | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .......................................................................TTTGTTGTTAGGTAAGCGTAT............................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................CAAAGCGACTCAAGCAGCT................. | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 |
| ...........................................................................TTGTTACGAAAGCGAATTCAA........................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................ATTCAAAGTTGGCATGATG.............................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................TTGGTTGCATAGTTAGCAA............................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................CATCGAAAAGCGTCGCACG...................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................GCACGCACGCAGCTCACCG............ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................AACATAGAAACGCGGCGCA........................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| dp5 | XL_group1a:8229135-8229385 + | dps_2579 | CATGGCAAGGGCGCCTTTCGCATGGAGGACTGCTTCATAACCGCCACAAAGTGGGTCAAGTGTGAAGTCCAAAACAACAATCCATTCGCATAAGTTTCCACTGTAGTACAACACAGTCTGGTTGCCAATGCACTAGAAATCAACATATCAATCGATCAATAACAATTCTCAAAT--ACACACATACACAATGCAATGAAAGTGTGTAAGTTTGTTTCTTTTCGCTGCGTGCGTCGAGTGGATTGTTGATCAGT |
| droPer2 | scaffold_15:589364-589616 + | CACGGCAAGAGCGCCTTTCGCATGGAGGACTGCTTCATAACCGCCACAAAGTGGGTCAAGTGTGAAGTCCAAAACAACAATCCATTCGCATAAGTTTACACTGTAGCACAACACTGTCTGGTTGCCAATGCACTAGAAATCAACATATCAATCGATCAAGAACAATTCTCAAATACACACACATTCACAAAGCAATGAAAGTGTGTAAGTTTGTTTCTTTTCGCTGCGTGCATCGAGTGGATTGTTGATCAGT | |
| droWil2 | scf2_1100000004515:1984028-1984070 - | CAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--CACACACACACGCAATACTGTGTAAGTATATATGTGTGT--------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/18/2015 at 04:56 AM